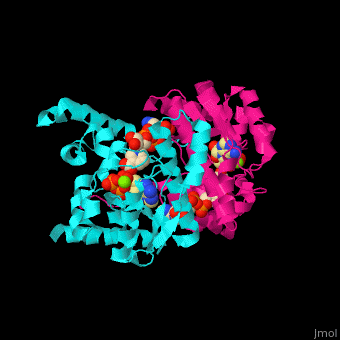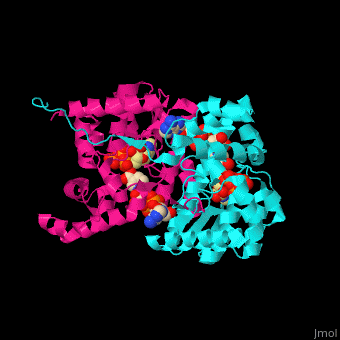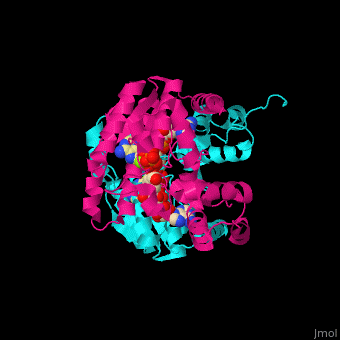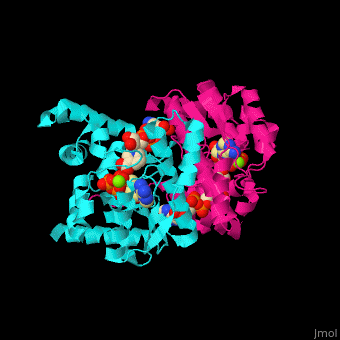We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
NAD synthase
From Proteopedia
(Difference between revisions)
| (21 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='' size='350' side='right' caption='NAD+ synthase complex with Mg+2 (green), ATP and deamino-NAD (PDB entry [[1xng]])' scene='54/541072/Cv/1'> | |
| - | + | ||
| - | + | ||
| + | == Function == | ||
| + | '''NAD+ synthase''' or '''NH3-dependent NAD synthetase''' (NADS) catalyzes the conversion of ATP, deamino-NAD+ and NH<sub>3</sub> to AMP, NAD+ and diphosphate (PP). NADS is part of the NAD synthesis pathway<ref>PMID:12898714</ref> . See [[NAD]]. | ||
| + | == Structural highlights == | ||
| + | The <scene name='54/541072/Cv/8'>ATP binding site</scene> contains <scene name='54/541072/Cv/9'>Mg+2 ion</scene>. The <scene name='54/541072/Cv/10'>NAD binding site is located in the interface of the 2 subunits</scene><ref>PMID:15645437</ref>. Water molecules are shown as red spheres. | ||
| + | </StructureSection> | ||
| + | == 3D Structures of NAD+ synthase == | ||
| + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| + | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| + | *'''NAD synthase''' | ||
| + | **[[3p52]] – NADS – ''Campylobacter jejuni'' <br /> | ||
| + | **[[3q4g]] – NADS (mutant) – ''Vibrio cholerae'' <br /> | ||
| + | **[[1wxf]] – EcNADS – ''Escherichia coli'' <br /> | ||
| + | **[[1xnh]] – HpNADS - ''Helicobacter pylori'' <br /> | ||
| + | **[[2pzb]] – BaNADS - ''Bacillus anthracis'' <br /> | ||
| + | **[[3dpi]] – NADS – ''Burkholderia pseudomallei''<br /> | ||
| + | **[[6kv3]] – NADS – ''Staphylococcus aureus''<br /> | ||
| + | **[[5wp0]] – NADS – ''Allivibrio fischeri''<br /> | ||
| + | **[[5huh]], [[5huj]] – NADS – ''Streptococcus pyogenes''<br /> | ||
| + | **[[3n05]] – NADS – ''Streptomyces avermitillis''<br /> | ||
| + | **[[4xfd]] – PaNADS – ''Pseudomonas aeruginosa''<br /> | ||
| + | **[[4q16]] – NADS – ''Deinococcus radiodurans''<br /> | ||
| + | **[[2e18]] – NADS – ''Pyrococcus horikoshii''<br /> | ||
| + | *NADS binary complex | ||
| + | **[[1ifx]] – BsNADS + deamino-NAD – ''Bacillus subtilis'' <br /> | ||
| + | **[[1wxh]] – EcNADS + NAD <br /> | ||
| + | **[[3hmq]] – BsNADS + NAD – ''Salmonella typhimurium'' <br /> | ||
| + | **[[6c8q]] – NADS + NAD – ''Enterococcus faecalis''<br /> | ||
| + | **[[5f23]] – PaNADS + NAD <br /> | ||
| + | *NADS ternary complex | ||
| + | **[[1ih8]] – BsNADS + Mg + ATP derivative <br /> | ||
| + | **[[1wxe]] – EcNADS + Mg + AMP <br /> | ||
| + | **[[1wxg]] – EcNADS + Mg + deamino-NAD <br /> | ||
| + | **[[2pz8]] – BaNADS + Mg + ATP derivative <br /> | ||
| + | **[[3fiu]] – NADS + Mg + AMP – ''Francisella tularensis''<br /> | ||
| + | *NADS quaternary complex | ||
| + | **[[1ee1]] – BsNADS + Mg + ATP + deamino-NAD <br /> | ||
| + | **[[1fyd]] – BsNADS + Mg + AMP + PP <br /> | ||
| + | **[[1kqp]] – BsNADS + Mg + PP + NAD derivative<br /> | ||
| + | **[[1wxi]] – EcNADS + Mg + AMP + PP <br /> | ||
| + | **[[1xng]] – HpNADS + Mg + ATP + deamino-NAD <br /> | ||
| + | **[[2pza]] – BaNADS + Mg + AMP + PP <br /> | ||
| + | *NADS quinary complex | ||
| + | **[[1nsy]] – BsNADS + Mg + ATP + ADP + PP <br /> | ||
| + | **[[2nsy]] – BsNADS + Mg + AMP + NAD + PP <br /> | ||
| + | *NADS glutamine-dependent | ||
| + | **[[6ofb]] – hNADS + AMP + NAD derivative<br /> | ||
| + | **[[3sdb]] – MtNADS (mutant) – ''Mycobacterium tuberculosis'' <br /> | ||
| + | **[[3seq]], [[3sez]] – MtNADS (mutant) + ATP derivative + pyridinium derivative <br /> | ||
| + | **[[3szg]] – MtNADS (mutant) + AMP + pyridinium derivative <br /> | ||
| + | **[[3syt]] – MtNADS + AMP + NAD + glutamate <br /> | ||
| + | **[[3dla]] – MtNADS + norleucine + deamido-NAD <br /> | ||
| + | **[[6ofc]] – MtNADS (mutant) + AMP + sulfonamide derivative + glutamate <br /> | ||
| + | **[[4f4h]] – NADS – ''Burkholderia thailandensis''<br /> | ||
| + | **[[3ilv]] – NADS – ''Cytophaga hutchinsonii''<br /> | ||
| - | + | }} | |
| - | + | == References == | |
| - | + | <references/> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | === | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
3D Structures of NAD+ synthase
Updated on 15-November-2021
References
- ↑ Suda Y, Tachikawa H, Yokota A, Nakanishi H, Yamashita N, Miura Y, Takahashi N. Saccharomyces cerevisiae QNS1 codes for NAD(+) synthetase that is functionally conserved in mammals. Yeast. 2003 Aug;20(11):995-1005. PMID:12898714 doi:http://dx.doi.org/10.1002/yea.1008
- ↑ Kang GB, Kim YS, Im YJ, Rho SH, Lee JH, Eom SH. Crystal structure of NH3-dependent NAD+ synthetase from Helicobacter pylori. Proteins. 2005 Mar 1;58(4):985-8. PMID:15645437 doi:10.1002/prot.20377