Purine repressor
From Proteopedia
(Difference between revisions)
| (16 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load=' | + | <StructureSection load='' size='450' side='right' caption='Structure of E. coli purine processor complex with DNA and guanine (PDB entry [[1wet]])' scene='55/554904/Cv/1'> |
| + | == Function == | ||
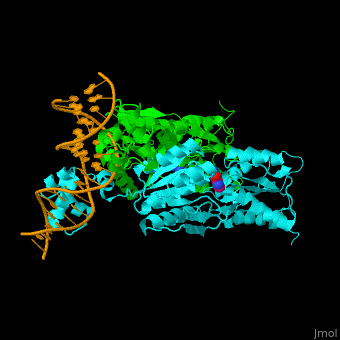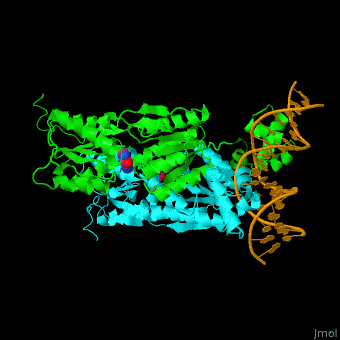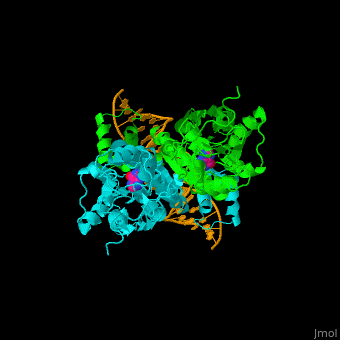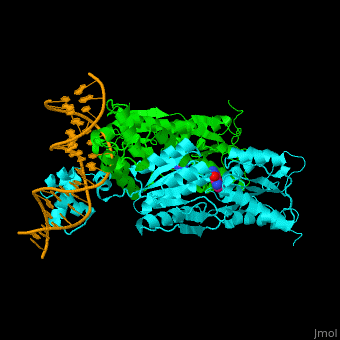
| + | '''Purine repressor''' (PurR) is a member of the lac repressor family. PurP binds DNA via a highly conserved helix-turn-helix at the N terminal (DBD). PurP contains a nucleotide co-repressor binding domain as well (CBD). PurP binds to a 16-bp operator sequence and co-regulates genes which are involved in the biosynthesis of purine and pyrimidine nucleotides<ref>PMID:1971621</ref>. | ||
| - | '' | + | == Structural highlights == |
| + | The <scene name='55/554904/Cv/5'>PurP guanine co-repressor binding site includes stacking interactions as well as hydrogen bonded ones</scene>. Water molecules are shown as red spheres. The DNA binding domain contains a <scene name='55/554904/Cv/6'>helix-turn-helix-loop-helix motif which interacts with the DNA major groove</scene> and a hinge helix binding to to the DNA minor groove. <scene name='55/554904/Cv/7'>Residue L54 interdigitates with the DNA central base pair</scene><ref>PMID:9278422</ref>. | ||
| + | </StructureSection> | ||
==3D structures of purine repressor== | ==3D structures of purine repressor== | ||
Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| + | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| - | + | *Purine repressor | |
| - | + | ||
| - | + | ||
| - | ''' | + | **[[1pru]], [[1prv]] - EcPurR DBD – ''Escherichia coli'' - NMR<br /> |
| + | **[[1dbq]] - EcPurR CBD + Mg - NMR<br /> | ||
| + | **[[1jhz]] - EcPurR CBD (mutant) + Mg <br /> | ||
| + | **[[1o57]] - BsPurR - ''Bacillus subtilis''<br /> | ||
| - | + | *Purine repressor complex with DNA and nucleotide | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | **[[1pnr]], [[1bdi]], [[1qp0]], [[1qp4]], [[1qpz]], [[1qqb]] - EcPurR + DNA + hypoxanthine <br /> | ||
| + | **[[1bdh]], [[2pud]], [[2pug]], [[1vpw]], [[1qp7]], [[1qqa]], [[1jfs]], [[1jft]], [[1jh9]] - EcPurR (mutant) + DNA + hypoxanthine <br /> | ||
| + | **[[1wet]] - EcPurR + DNA + guanine <br /> | ||
| + | **[[2pua]] - EcPurR (mutant) + DNA + methylpurine <br /> | ||
| + | **[[2pub]], [[2pue]] - EcPurR (mutant) + DNA + adenine <br /> | ||
| + | **[[2puc]], [[2puf]] - EcPurR (mutant) + DNA + guanine <br /> | ||
| + | **[[1zay]] - EcPurR + DNA + hypoxanthine + di-imino-purine<br /> | ||
| + | **[[7rmw]] – BsPurR + PPGPP<br /> | ||
| + | }} | ||
| + | == References == | ||
| + | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
3D structures of purine repressor
Updated on 08-February-2022
References
- ↑ Wilson HR, Turnbough CL Jr. Role of the purine repressor in the regulation of pyrimidine gene expression in Escherichia coli K-12. J Bacteriol. 1990 Jun;172(6):3208-13. PMID:1971621
- ↑ Schumacher MA, Glasfeld A, Zalkin H, Brennan RG. The X-ray structure of the PurR-guanine-purF operator complex reveals the contributions of complementary electrostatic surfaces and a water-mediated hydrogen bond to corepressor specificity and binding affinity. J Biol Chem. 1997 Sep 5;272(36):22648-53. PMID:9278422