We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox 124
From Proteopedia
(Difference between revisions)
| (46 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | <StructureSection load= size=550 side='right' scene='37/372724/Tp_1_in_grey/1'> | |
==='''Introduction'''=== | ==='''Introduction'''=== | ||
| - | + | Peptidoglycan transpeptidases, also known as penicillin-binding proteins | |
| - | catalyze the cross-linking of peptidoglycan polymers during bacterial cell wall | + | (PBP), catalyze the cross-linking of peptidoglycan polymers during |
| - | synthesis | + | bacterial cell wall synthesis. Beta-lactam (β-lactam) antibiotics, |
| - | + | which include penicillins, cephalosporins and carbapenems, bind and | |
| - | include penicillins, cephalosporins and carbapenems, bind and irreversibly | + | irreversibly inhibit transpeptidases. The overuse and misuse of β-lactam |
| - | inhibit transpeptidases | + | antibiotics has led to strains of Staphylococcus aureus that are resistant |
| - | + | to all β-lactams and are often only susceptible to “last resort antibiotics”, | |
| - | + | such as vancomycin. | |
| - | Staphylococcus aureus | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | antibiotics”, such as vancomycin. | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | ===''' | + | |
| - | PBP2a is composed of two domains: | + | ==='''Cell Wall Structure'''=== |
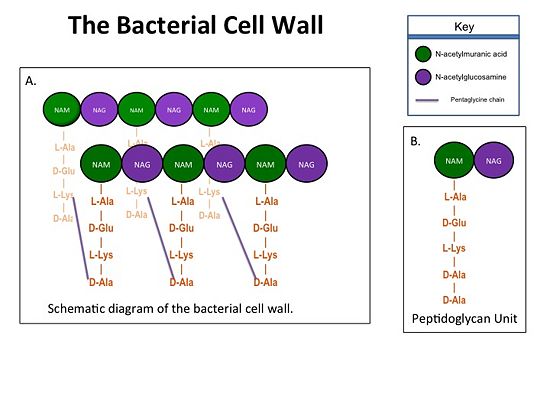
| - | <scene name='37/372724/Transpeptidase_domain/1'>TP</scene> domain. The NBP domain of PBP2a is anchored in the cell membrane, while the TP domain “sits” in the periplasm with its active site facing the inner surface of the cell wall. The active site contains a | + | The cell wall, which is composed of peptidoglycans, is crucial for maintaining |
| - | serine residue at position 403 (<scene name='37/372724/Serine403label/2'> | + | the structural integrity of the bacterium. Peptidoglycans consists of |
| + | N-acetylmuramic Acid (NAM) and N-acetylglucosamine (NAG) polymers. Rows of | ||
| + | peptidoglycan are cross-linked together with pentaglycine chains. The NAM residues | ||
| + | have a five amino acid side chain that terminates with two D-Alanine (D-Ala) | ||
| + | residues. [[Image:Cell Wall 7 30 2013.jpg|thumb|alt= Alt text| Figure 1.(A)Rows of Peptidoglycans forming a Bacterial Cell Wall (B)Peptidoglycan with D-Ala-D-Ala substrate |550px]] | ||
| + | |||
| + | |||
| + | ==='''Structure of a Resistant Transpeptidase'''=== | ||
| + | Methicillin resistant Staphylococcus aureus (MRSA) is resistant to all β-lactams because it acquires an alternative PBP, PBP2a, that is not bound or inhibited by any β-lactams. PBP2a is composed of two domains: | ||
| + | <scene name='37/372724/Non_penicillin_binding/1'>non penicillin binding(NPB)</scene> domain and a <scene name='37/372724/Transpeptidase_domain/1'>Transpeptidase(TP)</scene> domain. The NBP domain of PBP2a is anchored in the cell membrane, while the TP domain “sits” in the periplasm with its active site facing the inner surface of the cell wall. The active site contains a serine residue at position 403 (<scene name='37/372724/Serine403label/2'>Ser403</scene>)which catalyzes the cross-linking of the peptidoglycan rows with pentaglycine cross-links. | ||
| + | |||
| + | |||
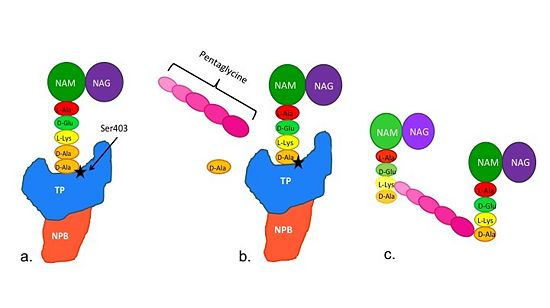
| + | ==='''Catalytic Mechanism of PBP2a'''=== | ||
| + | [[Image:Schematic TP 3steps.jpg|thumb|alt= Alt text|Figure 2. Schematic showing Catalytic Mechanism of PBP2a |550px]] | ||
| + | (a) The D-Ala-D-Ala side-chain substrate of the peptidoglycan accesses | ||
| + | the active site of the PBP2a. | ||
| + | |||
| + | (b) Ser403 nucleophilically attacks the peptide bond of the terminal | ||
| + | D-Ala residues of the substrate. The terminal D-Ala residue then exits | ||
| + | the active site. The now terminal D-Ala residue forms a covalent bond to Ser403, | ||
| + | while a crosslinking pentaglycine chain enters the active site. | ||
| + | |||
| + | (c) A covalent bond forms between the pentaglycine chain and the | ||
| + | terminal D-Ala residue, regenerating the active site serine residue. | ||
| + | |||
| + | The entire process takes 4 milliseconds. | ||
| + | |||
| + | ==='''How Do Antibiotics Work?'''=== | ||
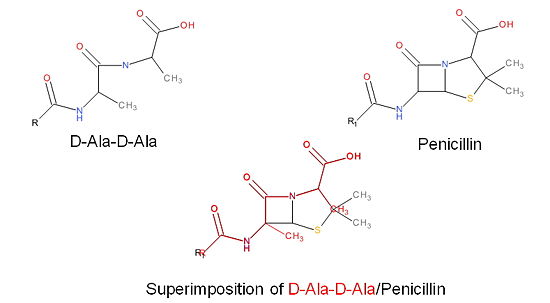
| + | The β-lactam antibiotics inhibit bacterial growth by inhibiting PBPs and ultimately cell wall | ||
| + | synthesis. Specifically, β-lactams are molecular mimics of D-Ala-D-Ala, which is the normal | ||
| + | substrate of PBPs. Nucleophillic attack of the β-lactam results in the PBP being irreversibly | ||
| + | inhibited by the β-lactam. As a result, the synthesis of the cell wall is inhibited which leads | ||
| + | to cell lysis. | ||
| + | [[Image:Structures on penicillin and b lactam.jpg|thumb|alt= Alt text|Figure 3. Mechanism of action of β-lactams. A. Structure of a β-lactam (penicillin) showing the amide, carboxyl, and β-lactam ring groups β-lactam ring groups. B. Structure of the D-Ala-D-Ala substrate. C. Overlay of the D-Ala-D-Ala substrate in red with penicillin demonstrating molecular mimicry.|550 px]] | ||
| + | |||
| + | |||
| + | ==='''PBP2a and Ceftobiprole'''=== | ||
| + | MRSA becomes resistant to β-lactams by acquiring an alternative PBP, PBP2a, that is | ||
| + | neither bound nor inhibited by β-lactams. Recently, two cephalosporins – | ||
| + | <scene name='37/372724/Ceftobiprole/1'>ceftobiprole</scene> and | ||
| + | ceftaroline – that have anti-MRSA activity have been developed. Ceftobiprole is able to | ||
| + | inhibit PBP2a because additional chemical groups at the | ||
| + | <scene name='37/372724/Ceftobiprole/7'>R2</scene> | ||
| + | position of the cephalosporin backbone are able to interact with additional amino acid | ||
| + | residues in PBP2a; specifically | ||
| + | <scene name='37/372724/Tyr446_and_met641_label/2'>Tyr446 and Met641</scene>. | ||
| + | As a result of ceftobiprole <scene name='37/372724/R2_interaction/4'>tighter binding</scene> to PBP2a as highlighted in green , <scene name='37/372724/Ceftobiprole_in_cpk/1'>the medicine</scene>, shown as colors of the atom types ([[CPK]]), is able to more efficiently react with the serine active site residue and therefore inhibit the activity of PBP2a. | ||
Current revision
| |||||||||||