This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Andrew Wills/Sandbox 1
From Proteopedia
(Difference between revisions)
| (5 intermediate revisions not shown.) | |||
| Line 15: | Line 15: | ||
==Key Structures== | ==Key Structures== | ||
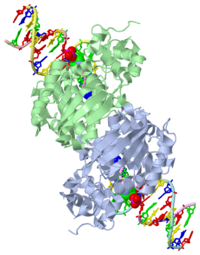
| - | AlkA is composed of three main domains with dimensions of approximately 50 Angstroms, 45 Angstroms, and 25 Angstroms.<ref name="Labahn">Labahn, Jorg, Orlando Scharer, et al. "Structural Basis for the Excision Repair of Alkylation-Damaged DNA." Cell. 86.2 (1996): 321-329. Print.</ref> The first domain (residues 1-112) is the <scene name='56/566536/N_terminal_domain_and_dna/ | + | AlkA is composed of three main domains with dimensions of approximately 50 Angstroms, 45 Angstroms, and 25 Angstroms.<ref name="Labahn">Labahn, Jorg, Orlando Scharer, et al. "Structural Basis for the Excision Repair of Alkylation-Damaged DNA." Cell. 86.2 (1996): 321-329. Print.</ref> The first domain (residues 1-112) is the <scene name='56/566536/N_terminal_domain_and_dna/4'>N-Terminal Domain</scene> that is composed of a five stranded antiparallel beta sheet and two alpha helices.<ref name="Labahn">Labahn, Jorg, Orlando Scharer, et al. "Structural Basis for the Excision Repair of Alkylation-Damaged DNA." Cell. 86.2 (1996): 321-329. Print.</ref> The <scene name='56/566536/Second_domain2/1'>Second Domain</scene> (residues 113-230) contains seven alpha helices that create a hydrophobic core.<ref name="Labahn">Labahn, Jorg, Orlando Scharer, et al. "Structural Basis for the Excision Repair of Alkylation-Damaged DNA." Cell. 86.2 (1996): 321-329. Print.</ref> The third domain (resudues 231-282) is the <scene name='56/566536/C_terminal_domain/2'>C-Terminal Domain</scene> that contains a bundle of three alpha helices. <ref name="Labahn">Labahn, Jorg, Orlando Scharer, et al. "Structural Basis for the Excision Repair of Alkylation-Damaged DNA." Cell. 86.2 (1996): 321-329. Print.</ref> The three domains of AlkA create a distinct binding structure that is lined with electron rich aromatic residues to form a hydrophobic cleft. The cleft is found between domains 2 and 3, and forms the binding pocket for the alkylated base. The hydrophobic cleft has the ability to widen between domains 2 and 3 for accepting a variety of alkylated bases.<ref name="Labahn">Labahn, Jorg, Orlando Scharer, et al. "Structural Basis for the Excision Repair of Alkylation-Damaged DNA." Cell. 86.2 (1996): 321-329. Print.</ref> |
AlkA is a member of the helix-hairpin-helix (HhH) family of DNA glycosylases, where two compact alpha helical structures are connected by a hairpin loop.<ref name="Moe">Moe, E, D.R. Hall, et al. "Structure-function studies of an unusual 3-methyladenine DNA glycosylase II (AlkA) from Deinococcus radiodurans." Biological Crystallography. 68.6 (2012): 703-712. Print. </ref> In AlkA, the <scene name='56/566536/Helix_hairpin_helix_domain/3'>HhH Domain</scene> is located on the rim of the active site and is composed of residues 202-227.<ref name="Hollis">Hollis, Thomas, Yoshitaka Ichikawa, and Tom Ellenberger. "DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA." EMBO Journal. 19.4 (2000): 758-766. Print. </ref> The HhH sequence is responsible for binding the damaged DNA by van der Waals interactions, a few hydrogen bonds, and metal ion interactions. | AlkA is a member of the helix-hairpin-helix (HhH) family of DNA glycosylases, where two compact alpha helical structures are connected by a hairpin loop.<ref name="Moe">Moe, E, D.R. Hall, et al. "Structure-function studies of an unusual 3-methyladenine DNA glycosylase II (AlkA) from Deinococcus radiodurans." Biological Crystallography. 68.6 (2012): 703-712. Print. </ref> In AlkA, the <scene name='56/566536/Helix_hairpin_helix_domain/3'>HhH Domain</scene> is located on the rim of the active site and is composed of residues 202-227.<ref name="Hollis">Hollis, Thomas, Yoshitaka Ichikawa, and Tom Ellenberger. "DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA." EMBO Journal. 19.4 (2000): 758-766. Print. </ref> The HhH sequence is responsible for binding the damaged DNA by van der Waals interactions, a few hydrogen bonds, and metal ion interactions. | ||
| Line 25: | Line 25: | ||
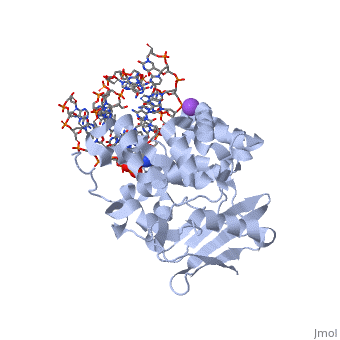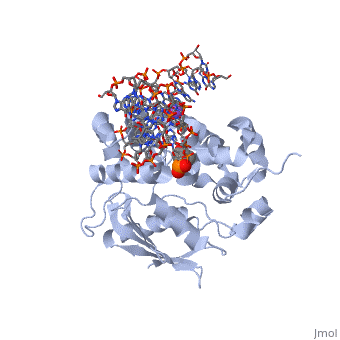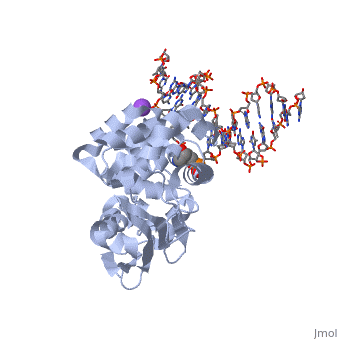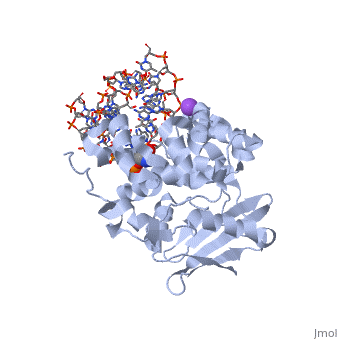
The AlkA protein stabilizes DNA by polar and nonpolar interactions with the DNA backbone. There are few positively charged residues on the AlkA’s binding surface that could interact with the DNA backbone, which is why the majority of polar interactions involve the DNA strand containing the alkylated base. | The AlkA protein stabilizes DNA by polar and nonpolar interactions with the DNA backbone. There are few positively charged residues on the AlkA’s binding surface that could interact with the DNA backbone, which is why the majority of polar interactions involve the DNA strand containing the alkylated base. | ||
| - | DNA is held in place by interactions with the HhH segment, which acts as a platform to create the 66 degree bend in DNA mentioned earlier. The bending of the DNA relieves tensional strain created by the widening of the minor groove for base flipping.<ref name="Hollis">Hollis, Thomas, Yoshitaka Ichikawa, and Tom Ellenberger. "DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA." EMBO Journal. 19.4 (2000): 758-766. Print. </ref> Amide nitrogens on residues Gly214, Gly216, and Thr219 <scene name='56/566536/Hydrogen_bonding/ | + | DNA is held in place by interactions with the HhH segment, which acts as a platform to create the 66 degree bend in DNA mentioned earlier. The bending of the DNA relieves tensional strain created by the widening of the minor groove for base flipping.<ref name="Hollis">Hollis, Thomas, Yoshitaka Ichikawa, and Tom Ellenberger. "DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA." EMBO Journal. 19.4 (2000): 758-766. Print. </ref> Amide nitrogens on residues Gly214, Gly216, and Thr219 <scene name='56/566536/Hydrogen_bonding/5'>hydrogen bond</scene> with DNA’s phosphodiester backbone, while residues Gln210, Phe212, and Ile215 <scene name='56/566536/Coordinate_sodium_ion/7'>coordinate</scene> the sodium metal ion that interacts with DNA further stabilize it in the molecule.<ref name="Hollis">Hollis, Thomas, Yoshitaka Ichikawa, and Tom Ellenberger. "DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA." EMBO Journal. 19.4 (2000): 758-766. Print. </ref> |
In order to keep the DNA strand bound to the protein, AlkA depends on van der Waals interactions on the minor groove of DNA. The van der Waals interactions play a major role in AlkA’s preference for double stranded DNA.<ref name="Hollis">Hollis, Thomas, Yoshitaka Ichikawa, and Tom Ellenberger. "DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA." EMBO Journal. 19.4 (2000): 758-766. Print. </ref> An example of this may be seen with the Pro175 wedged into the minor groove of DNA and anchoring it by van der Waals interctions.<ref name="Hollis">Hollis, Thomas, Yoshitaka Ichikawa, and Tom Ellenberger. "DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA." EMBO Journal. 19.4 (2000): 758-766. Print. </ref> | In order to keep the DNA strand bound to the protein, AlkA depends on van der Waals interactions on the minor groove of DNA. The van der Waals interactions play a major role in AlkA’s preference for double stranded DNA.<ref name="Hollis">Hollis, Thomas, Yoshitaka Ichikawa, and Tom Ellenberger. "DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA." EMBO Journal. 19.4 (2000): 758-766. Print. </ref> An example of this may be seen with the Pro175 wedged into the minor groove of DNA and anchoring it by van der Waals interctions.<ref name="Hollis">Hollis, Thomas, Yoshitaka Ichikawa, and Tom Ellenberger. "DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA." EMBO Journal. 19.4 (2000): 758-766. Print. </ref> | ||
Current revision
| |||||||||
| 1diz, resolution 2.50Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | |||||||||
| Non-Standard Residues: | |||||||||
| Activity: | DNA-3-methyladenine glycosylase II, with EC number 3.2.2.21 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
CRYSTAL STRUCTURE OF E. COLI 3-METHYLADENINE DNA GLYCOSYLASE (ALKA) COMPLEXED WITH DNA
| |||||||||||
See Also
References
- ↑ 1.0 1.1 Berg, Jeremy, Tymoczko John, and Lubert Stryer. Biochemistry. 6th. New York: W.H. Freeman and Company, 2007. 806-808. Print.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 Hollis, Thomas, Yoshitaka Ichikawa, and Tom Ellenberger. "DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA." EMBO Journal. 19.4 (2000): 758-766. Print.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 Labahn, Jorg, Orlando Scharer, et al. "Structural Basis for the Excision Repair of Alkylation-Damaged DNA." Cell. 86.2 (1996): 321-329. Print.
- ↑ 4.0 4.1 4.2 4.3 Moe, E, D.R. Hall, et al. "Structure-function studies of an unusual 3-methyladenine DNA glycosylase II (AlkA) from Deinococcus radiodurans." Biological Crystallography. 68.6 (2012): 703-712. Print.