We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Single stranded binding protein
From Proteopedia
(Difference between revisions)
(→Structure of ''E. coli'' SSB) |
|||
| (3 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='1eyg' size='400' side='right' frame='true' caption='E. coli single-stranded DNA-binding protein chymotryptic fragment complex with DNA (PDB code [[1eyg]])' scene=''> | |
| + | Single Stranded DNA-Binding Protein (SSB) | ||
==Overview== | ==Overview== | ||
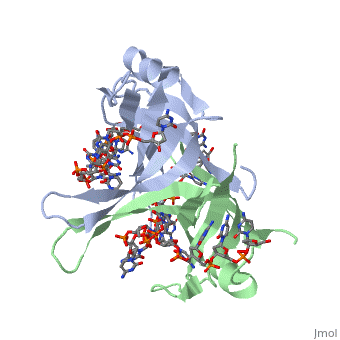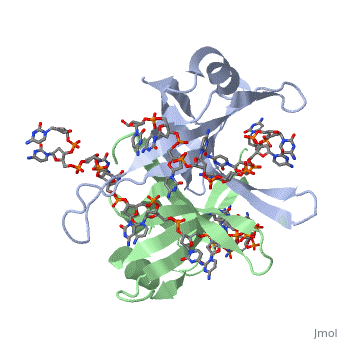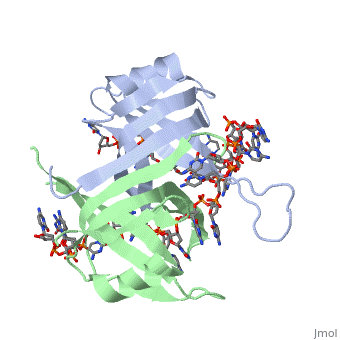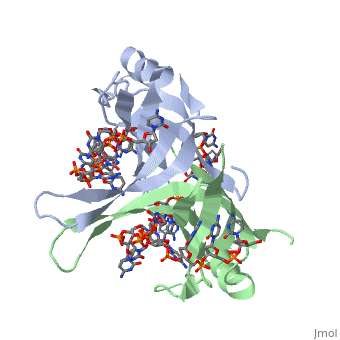
'''Single-stranded DNA-binding protein''' '''(SSB)''' binds to single-stranded regions of DNA. This binding serves a variety of functions - it prevents the strands from hardening too early during replication, it protects the single-stranded DNA from being broken down by nucleases during repair, and it removes the secondary structure of the strands so that other enzymes are able to access them and act effectively upon the strands<ref>PMID:2087220</ref>. | '''Single-stranded DNA-binding protein''' '''(SSB)''' binds to single-stranded regions of DNA. This binding serves a variety of functions - it prevents the strands from hardening too early during replication, it protects the single-stranded DNA from being broken down by nucleases during repair, and it removes the secondary structure of the strands so that other enzymes are able to access them and act effectively upon the strands<ref>PMID:2087220</ref>. | ||
| Line 7: | Line 8: | ||
==Structure of ''E. coli'' SSB== | ==Structure of ''E. coli'' SSB== | ||
| - | + | ||
SSB proteins have been identified in many different organisms, but the most well understood SSB remains the SSB of ''E. coli''. ''E. coli'' SSB is a homotetramer consisting of <scene name='56/566528/Homotetramer/1'>four identical subunits</scene> which are each about 19 kDa in size <ref>PMID:2087220</ref>. There are two different binding modes of the ''E. coli'' SSB when it complexes with ssDNA<ref>PMID:11993998</ref>. Regulation of these modes has been found to be dependent on salt concentration, in addition to other unknown factors. Under low salt conditions, the protein is less efficient as only two of the four identical subunits of ''E. coli'' SSB were found to bind to the ssDNA <ref>PMID:11993998</ref>. This is a common theme among DNA binding proteins. The cause is presumed to be that the protein has less ion decoration at lower salt levels. And it could be that the subunits interact with each other through salt bridges to remain close to each other and the DNA. Under high salt concentrations, however, all four subunits of the homotetramer bind to the ssDNA, increasing the number of nucleotides in contact with the SSB and thus favoring SSB-ssDNA interactions. Depending on the salt concentration and other factors, estimates of the size of the site of interaction between SSB and ssDNA range anywhere from 30 to 73 nucleotides for each tetramer <ref>PMID:11993998</ref>. | SSB proteins have been identified in many different organisms, but the most well understood SSB remains the SSB of ''E. coli''. ''E. coli'' SSB is a homotetramer consisting of <scene name='56/566528/Homotetramer/1'>four identical subunits</scene> which are each about 19 kDa in size <ref>PMID:2087220</ref>. There are two different binding modes of the ''E. coli'' SSB when it complexes with ssDNA<ref>PMID:11993998</ref>. Regulation of these modes has been found to be dependent on salt concentration, in addition to other unknown factors. Under low salt conditions, the protein is less efficient as only two of the four identical subunits of ''E. coli'' SSB were found to bind to the ssDNA <ref>PMID:11993998</ref>. This is a common theme among DNA binding proteins. The cause is presumed to be that the protein has less ion decoration at lower salt levels. And it could be that the subunits interact with each other through salt bridges to remain close to each other and the DNA. Under high salt concentrations, however, all four subunits of the homotetramer bind to the ssDNA, increasing the number of nucleotides in contact with the SSB and thus favoring SSB-ssDNA interactions. Depending on the salt concentration and other factors, estimates of the size of the site of interaction between SSB and ssDNA range anywhere from 30 to 73 nucleotides for each tetramer <ref>PMID:11993998</ref>. | ||
| Line 25: | Line 26: | ||
<scene name='56/566528/Gly_15/2'>Gly15</scene> is believed to play an important role in binding the RecA protein. SSB will interact with the protein RecA to enable recombination, because RecA will recognize SSB and replace it on the strand. In DNA repair, SSB will bind to the damaged strand to protect it. And eventually it will attract repair enzymes which will replace SSB and begin repair mechanisms. Mutations in <scene name='56/566528/Gly_15/2'>Gly15</scene> have been shown to have extreme effects on recombinational repair. SSB is also thought to bind with exonuclease I, DNA polymerase II, and a protein n, which is a part of the primosome complex and used to help synthesize RNA primers for the lagging strand <ref>PMID: 2087220</ref>. | <scene name='56/566528/Gly_15/2'>Gly15</scene> is believed to play an important role in binding the RecA protein. SSB will interact with the protein RecA to enable recombination, because RecA will recognize SSB and replace it on the strand. In DNA repair, SSB will bind to the damaged strand to protect it. And eventually it will attract repair enzymes which will replace SSB and begin repair mechanisms. Mutations in <scene name='56/566528/Gly_15/2'>Gly15</scene> have been shown to have extreme effects on recombinational repair. SSB is also thought to bind with exonuclease I, DNA polymerase II, and a protein n, which is a part of the primosome complex and used to help synthesize RNA primers for the lagging strand <ref>PMID: 2087220</ref>. | ||
| - | |||
| Line 34: | Line 34: | ||
As single-stranded DNA binding proteins are utilized in some of the most important aspects of DNA metabolism, they are used extensively in DNA replication, repair and recombination<ref>PMID: 2087220</ref>. Most SSBs use one or more subunits with an OB-fold motif to bind securely and preferentially to ssDNA. A few specific SSBs (such as RecA and adenovirus DBP) do not use the OB-fold, instead relying on electrostatic and stacking interactions as well as hydrogen bonding<ref>Shamoo, Yousif. “Single Stranded DNA binding proteins.” ‘’Encyclopedia of Life Sciences.’’ MacMillan Publishers Ltd, Nature Publishing Group; 2002</ref>. | As single-stranded DNA binding proteins are utilized in some of the most important aspects of DNA metabolism, they are used extensively in DNA replication, repair and recombination<ref>PMID: 2087220</ref>. Most SSBs use one or more subunits with an OB-fold motif to bind securely and preferentially to ssDNA. A few specific SSBs (such as RecA and adenovirus DBP) do not use the OB-fold, instead relying on electrostatic and stacking interactions as well as hydrogen bonding<ref>Shamoo, Yousif. “Single Stranded DNA binding proteins.” ‘’Encyclopedia of Life Sciences.’’ MacMillan Publishers Ltd, Nature Publishing Group; 2002</ref>. | ||
| - | + | ||
==See Also== | ==See Also== | ||
Current revision
| |||||||||||
Proteopedia Page Contributors and Editors (what is this?)
Refayat Ahsen, Rachel Craig, Michal Harel, Alexander Berchansky