Sandbox Reserved 779
From Proteopedia
| (83 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | [[Image:Example.jpg<!-- PLEASE DO NOT DELETE THIS TEMPLATE --> | + | [[Image:Example.jpg]<!-- PLEASE DO NOT DELETE THIS TEMPLATE --> |
{{User:Michael_B._Goshe/Template_BCH455_555}} | {{User:Michael_B._Goshe/Template_BCH455_555}} | ||
<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | <!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
| + | '''β-Lactoglobulin''' | ||
| + | <Structure load='1beb' size='320' frame='true' align='left' caption='3D model 1._The dimer interface of β-lactoglobulin lattice X_1BEB' scene='Insert optional scene name here' /> | ||
| + | =='''β-Lactoglobulin'''== | ||
| - | < | + | β-Lactoglobulin (β-LG) was first isolated in 1934.<ref>http://www.jbc.org/content/104/2/359.citation</ref> It is the main globular protein of whey, a by-product from cow milk-cheese manufacture. Cow milk itself contains 20% whey proteins and 80% casein protein. β-Lactoglobulin constitutes 50-65% dry solids whey protein or 12% of whole cow milk proteins. |
| - | ---- | + | Due to its abundance, and relatively easy to isolate nature, β-Lactoglobulin used widely in Industry to increase the protein contents of the food and beverage products. |
| + | Bovine β-lactoglobulin (β-Lg) is a commercially important whey protein with undetermined biological function, although it is of obvious nutritional value. | ||
| + | β-Lg binds a variety of ligands, and it appears that there are at least 3 independent binding sites: calyx, putative grove, and dimer interface (Fig. 2).<ref>http://www.sciencedirect.com/science/article/pii/S0958694698000211</ref><ref>PMID:9867826</ref> | ||
| + | β-Lactoglobulin amino-acid sequence and 3-dimensional structure show that it belongs to [[Lipocalin]] family which capable of binding hydrophobic ligands and thus may act as specific transporters, as does serum retinol binding protein. <ref>PMID:15259212</ref> Bovine β-Lactoglobulin is synthesized in cow mammary gland and secreted in the milk. It causes an allergic reaction in human and is one of the causes of cow's milk allergy. | ||
| - | + | ===Lipocalin Proteins=== | |
| + | β-Lactoglobulin belongs to the calycin superfamily and Lipocalin family. Lipocalins are typically small (160-180 residues in length), extracellular proteins and able to bind small hydrophobic molecules (such as retinol); bind to specific cell-surface receptors; and form of covalent and non-covalent complexes with other soluble macromolecules. Lipocalin proteins have also been classified mainly as transport proteins. | ||
| + | <ref>PMID:11058743</ref> | ||
| - | + | The lipocalin family is a large and diverse family of proteins with functions varying from insect camouflage to small hydrophobic molecule transport typified by the serum retinol-binding protein <ref>http://www.biochemj.org/bj/318/bj3180001.htm</ref> The crystal structures so far determined reveal the typical lipocalin to be an eight-stranded antiparallel β-barrel arranged to form a conical central calyx or cavity in which the hydrophobic ligand is located.<ref>PMID:6540172</ref> | |
| + | [[Image:Beta_lac_Residues.jpg|thumb|right|320px|Figure 1. Primary structure of β-LG. β-Lactoglobulin comprises 162 AA, including 5 Cys residues. Two disulfide linkages are located at residues Cys-106 to Cys-119 and Cys-66 to Cys-160. One free Cys is at position 121.(Taken from Liu, et al. 2007)<ref>PMID:17235131</ref>]] | ||
| + | [[Image:General_view_of_beta_lactoglobulin.jpg|thumb|left|320px|Figure 2. A general view of b-lactoglobulin, a typical lipocalin. The binding site (filled atoms) is shown in the central calyx, and the putative binding site (open atoms) is indicated on the outer surface of the protein. The structurally conserved regions are at the rear of the molecule on strand A, the FG loop, and the loop before the a-helix.(Wu S et al. J. Biol. Chem. 1999;274:170-174)<ref>PMID:9867826</ref>]] | ||
| - | + | =='''Structure of β-Lactoglobulin'''== | |
| + | β-Lactoglobulin is a small globulin protein, soluble in dilute salt solution with 162 amino acid residues (Mr ∼18,400 Daltons) for each monomer that fold up into an 8-stranded, antiparallel β-barrel with a 3-turn α-helix on the outer surface and a ninth β-strand flanking the first strand. | ||
| - | + | ===Residues and secondary structures=== | |
| + | β-Lactoglobulin contains two disulfide bonds (Cys 66–Cys 160 and Cys 106–Cys 119) and a free thiol (Cys 121)(Fig. 1). Structures of βLG have been reported by several groups with X-ray crystallography and solution NMR, that it is predominantly β-sheet protein. The β-barrel, or so called calyx, is conical and is made of two β-sheets: the B–D strands and N-terminal half of the A strand (denoted AN) form one sheet, and the E–H strands and C-terminal half of the A strand (denoted AC) form the other. On the outer surface of the β-barrel, between the G and H strands, is the 3-turn α-helix. The loops that connect the β-strands at the closed end of the calyx, BC, DE, and FG, are generally quite short, whereas those at the open end, AB, CD, EF,and GH, are significantly longer and more flexible. In the calyx, there is a large central cavity which is surrounded by hydrophobic residues and is accessible to solvent. This cavity provides the principal ligand-binding site (Fig. 2). <ref>PMID:19362581</ref> | ||
| - | + | ===Genetic Variants=== | |
| - | + | Genetically, β-lactoglobulin may exist as one of several variants, among which the variants A and B are the most abundant. The A and B variants of the protein differ from each other by amino acid residues at positions Asp64 (Gly64 in variant B) and Val118 (Ala118 in variant B). These differences in primary structure render the two variants slightly different with respect to isoelectric point, solubility, self-association properties, as well as pressure and temperature stability. However, the structural characteristics of the A and B variants of bovine b-lactoglobulin are virtually indistinguishable. In its native state, β-lactoglobulin is a predominantly β-sheet protein containing nine b-strands and three a-helices. The core of the protein is formed by a flattened b-barrel (a calyx) composed of eight antiparallel b-strands (A to H).<ref>PMID:11734004</ref> | |
| - | + | [[Image:BLG_Dimer_1BEB_Chain_A&B.png|thumb|right|240px|Figure 3. Bovine β-Lactoglobulin Dimer_[[1BEB]]]] | |
| + | ===Dimer/Monomer=== | ||
| + | At physiological conditions, majority of bovine b-lactoglobulin forms a dimer (Fig. 3). Below pH 3, the dimer dissociates into monomers which maintain their native conformation. | ||
| + | Dimeric Lactoglobulin molecules exist in the open conformation at basic pH, whereas they exist in the closed conformation at acidic pH, after undergoing Tanford transition around neutral pH.<ref>PMID:17932936</ref> | ||
| - | + | ===Ligands and Active sites=== | |
| - | + | The true function of β-Lg is unknown, but it has been suggested that it is involved in the transport of retinol and/or fatty acids. It binds retinol with a higher affinity than does RBP. β-Lg contains two tryptophans, Trp 19 on the A strand and Trp 61 on the C strand. The former is buried in the hydrophobic core whereas the latter is exposed to the solvent in the native structure, making them useful probes for monitoring site-specific conformational changes<ref>PMID:19362581</ref> and their fluorescence is altered when retinol is bound.<ref>PMID:9115437</ref> | |
| - | + | Co-crystallized β-Lg with palmitic acid (3D Model 2._1B0O) and the refined structure reveals that the ligand binds in the central cavity in a manner similar to the binding of retinol to the related lipocalin, serum retinol-binding protein. The carboxyl group binds to both Lys-60 and Lys-69 at the entrance to the cavity. The hydrophobic tail stretches in an almost fully extended conformation into the center of the protein.<ref>PMID:9867826</ref> | |
| - | + | In addition, studies on the monomer–dimer equilibrium and the reactivity of the thiol group of Cys121 which deeply buried between the α-helix and H strand revealed other important properties of β-LG. The stability of the structure also depend so heavily upon the external loop around residue 64 or the β strand with the free thiol.<ref>PMID:9115437</ref> | |
| + | [[Image:Vitamin D3 binding to the beta-lac calyx and dimer interface.jpg|thumb|left|240px|Figure 4. Vitamin D3 binding to the beta-lac calyx and dimer interface (Dominguez-Ramirez et al, 2013)<ref>PMID:24255705</ref>]] | ||
| + | β-Lg has binding sites for hydrophobic ligands like fatty acids, retinoic acid, and Vitamin D3 (cholecalciferol) and lactose [[ligands]] <ref>PMID:24255705</ref> For hydrophobic ligands two sites have been postulated, one inside the calyx and the other at the dimer interface, on the outer surface of the protein between the α-helix and the β-barrel (Fig. 4). The accessibility to the calyx is pH-dependent. | ||
| + | NMR and Xray analysis showed that the access is mediated by the mobile EF loop. All the structures with ligands bound to the calyx exhibit an open EF loop, suggesting that this site is accessible at neutral pH. | ||
| + | <Structure load='1b0o' size='320' frame='true' align='right' caption='3D model 2._β-lactoglobulin complexed with Palmitate, lattice Z_1B0O' scene='Insert optional scene name here' /> | ||
| - | + | ====Biological function==== | |
| - | + | Besides as binding protein, β-Lg is also assumed to have a transporter function especially for several ligands through the digestive tract in, as the calyx can protect any ligands entered/bound to the cavity.<ref>PMID:15259212</ref>. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | =='''Molecular mechanism of the Tanford transition'''== | ||
| - | + | The Tanford transition is a conformational change of bovine β-lactoglobulin occurring at around pH 7, identified originally on the basis of optical rotatory dispersion and the accessibility of a thiol group. X-ray analysis has suggested that a conformational change to the EF-loop (residues I84 -N90) (Fig. 5) is responsible for the Tanford transition, with the loop closing the hydrophobic cavity of the β-barrel of the β-LG molecule below pH 7 and flipping to open the cavity above pH 7.<ref>PMID:16368109</ref>Tanford transition is triggered by protonation of Glu89 exhibiting an anomalously high pKa value. | |
| - | The | + | The Tanford transition may involve some other structural changes as well. For example, the transition is accompanied by a change in the microenvironment of Tyr42 and causes an alteration in the relative orientation of monomers in the dimer by as much as 5 degrees, which breaks a number of intersubunit hydrogen bonds. <ref>PMID:11734004</ref> |
| - | + | [[Image:BLG_monomer_showing_EF_Loop.png|thumb|left|240px|Figure 5. Bovine β-Lactoglobulin Monomer showing EF loop (colored yellow)]] | |
| - | + | ||
| - | + | ||
| + | The structures of the trigonal crystal form of bovine β-lactoglobulin variant A at pH 6.2, 7.1, and 8.2 have been determined by X-ray diffraction methods. The glutamate side chain of residue 89 is buried at pH 6.2 and becomes exposed at pH 7.1 and 8.2. This conformational change, involving the loop 84-90, provides a structural basis for a variety of pH-dependent chemical, physical, and spectroscopic phenomena, collectively known as the Tanford transition.<ref>PMID:9760236</ref>It was found that the stereochemical environment of Tyr42 changes significantly with pH variation between pH 7 and pH 8. This may provide a structural explanation for an as yet unexplained feature of the Tanford transition, namely the increase in exposure of a tyrosine residue.<ref>PMID:11168385</ref> | ||
| - | == | + | =='''Uses / Application'''== |
| - | + | ===Food Industry and Pharmacy=== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | β-Lactoglobulin interaction with hydrophobic molecules and with other proteins, and its sensitivity to chemical, thermal and baric denaturation, all with a view to establishing relationships among structure, properties and functionality and its potential use in food and pharmacy industry.<ref>http://www.sciencedirect.com/science/article/pii/S0958694698000211</ref> | ||
| + | Food Industry has been widely utilize β-LG to increase functionality (sensory, texture, nutritional) in commercial food and beverages application, and also as inexpensive source of protein to increase the overall protein content of their product. Pharmacy Industry is recently investigating and developing design of drugs that utilize β-LG as vehicle to transport antioxidants, drugs and vitamins to the gut.<ref>http://www.faqs.org/patents/app/20110038942</ref> | ||
| - | + | ===Antioxidant Nature=== | |
| - | + | In the dairy industry, bovine milk is frequently heated for pasteurization (62.5°C for 30 min) and sterilization. This heating process may induce oxidative losses of proteins, unsaturated lipids, vitamins, active enzymes, and immunological factors. Cross-linking the free thiol groups of β-LG by heating (100 °C for 2 min), or chemically modifying the β-LG by carboxymethylation to block the thiol groups resulted in a substantial loss of antioxidant activity. The data suggest that Cys-121 plays an essential role in the antioxidant nature of β-LG. Because β-LG is extremely sensitive to thermal denaturation, to maintain its antioxidant nature, dairy products consumed daily should not be overheated in order to maintain its antioxidant nature.<ref>PMID:17235131</ref> | |
| + | Whey Protein whose majority constituent is β-Lactoglobulin, gives abundant source of branched chain amino acids especially Leucine which plays a key role in initiating transcription of protein synthesis that speed recovery and adaptation to stress (exercise) and also a free thiol Cys which is (postulated) utilized in the body to generate antioxidant glutathione.<ref>http://www.kettlercise.com/members-area/supplements/whey-protein.html</ref> | ||
| - | protein fold and how thats important for the function | ||
| - | ligands if theres ligands | ||
| - | the active site if relevant | ||
| - | features of protein that are important for function | ||
| - | zoom in on the active site, label the important active site residues, and hughlight those residues in a different color (make it look pretty) | ||
| - | == | + | =='''Other β-Lactoglobulin related 3D structures and complexes'''== |
| - | + | ||
| - | + | ||
| - | + | [[2q2m]] - Bovine β-Lactoglobulin Native (Fig. 4) | |
| - | + | ||
| - | + | [[1b8e]] - Crystal structure of the Bovine β-Lactoglobulin (Isoforms A and B) in orthorombic space group | |
| - | + | ||
| + | [[1qg5]] - Crystal structure of the Bovine β-Lactoglobulin (Isoforms A) | ||
| + | [[1beb]] - Bovine β-Lactoglobulin, Lattice X | ||
| - | + | [[1cj5]] - Bovine β-Lactoglobulin A | |
| + | [[1gx8]] - Bovine β-Lactoglobulin complexed with Retinol, Trigonal Lattice Z | ||
| - | + | [[1gx9]] - Bovine β-Lactoglobulin complexed with Retinoic acid, Trigonal Lattice Z | |
| - | + | ||
| - | + | ||
| - | [[ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | [[ | + | [[1gxa]] - Bovine β-Lactoglobulin complexed with Retinol and Palmitic acid, Trigonal Lattice Z |
| + | [[1b0o]] - Bovine β-Lactoglobulin complexed with Palmitate, Lattice Z | ||
| + | [[1bsy]] [[2blg]] [[3blg]] - Structural Basis of the Tanford Transitioon of Bovine β-Lactoglobulin from crystal structures at 3 pH values | ||
| - | + | =='''References'''== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | = | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<references/> | <references/> | ||
| - | == Proteopedia Page Contributors and Editors == | + | =='''Proteopedia Page Contributors and Editors'''== |
[[User:Rini Triani|Rini Triani]] | [[User:Rini Triani|Rini Triani]] | ||
| - | |||
| - | |||
[[Category:Bos taurus]] | [[Category:Bos taurus]] | ||
[[Category:Lipocalin]] | [[Category:Lipocalin]] | ||
Current revision
[[Image:Example.jpg]
| This Sandbox is Reserved from Sep 25, 2013, through Mar 31, 2014 for use in the course "BCH455/555 Proteins and Molecular Mechanisms" taught by Michael B. Goshe at the North Carolina State University. This reservation includes Sandbox Reserved 299, Sandbox Reserved 300 and Sandbox Reserved 760 through Sandbox Reserved 779. |
To get started:
More help: Help:Editing |
β-Lactoglobulin
|
Contents |
β-Lactoglobulin
β-Lactoglobulin (β-LG) was first isolated in 1934.[1] It is the main globular protein of whey, a by-product from cow milk-cheese manufacture. Cow milk itself contains 20% whey proteins and 80% casein protein. β-Lactoglobulin constitutes 50-65% dry solids whey protein or 12% of whole cow milk proteins. Due to its abundance, and relatively easy to isolate nature, β-Lactoglobulin used widely in Industry to increase the protein contents of the food and beverage products. Bovine β-lactoglobulin (β-Lg) is a commercially important whey protein with undetermined biological function, although it is of obvious nutritional value. β-Lg binds a variety of ligands, and it appears that there are at least 3 independent binding sites: calyx, putative grove, and dimer interface (Fig. 2).[2][3] β-Lactoglobulin amino-acid sequence and 3-dimensional structure show that it belongs to Lipocalin family which capable of binding hydrophobic ligands and thus may act as specific transporters, as does serum retinol binding protein. [4] Bovine β-Lactoglobulin is synthesized in cow mammary gland and secreted in the milk. It causes an allergic reaction in human and is one of the causes of cow's milk allergy.
Lipocalin Proteins
β-Lactoglobulin belongs to the calycin superfamily and Lipocalin family. Lipocalins are typically small (160-180 residues in length), extracellular proteins and able to bind small hydrophobic molecules (such as retinol); bind to specific cell-surface receptors; and form of covalent and non-covalent complexes with other soluble macromolecules. Lipocalin proteins have also been classified mainly as transport proteins. [5]
The lipocalin family is a large and diverse family of proteins with functions varying from insect camouflage to small hydrophobic molecule transport typified by the serum retinol-binding protein [6] The crystal structures so far determined reveal the typical lipocalin to be an eight-stranded antiparallel β-barrel arranged to form a conical central calyx or cavity in which the hydrophobic ligand is located.[7]

Structure of β-Lactoglobulin
β-Lactoglobulin is a small globulin protein, soluble in dilute salt solution with 162 amino acid residues (Mr ∼18,400 Daltons) for each monomer that fold up into an 8-stranded, antiparallel β-barrel with a 3-turn α-helix on the outer surface and a ninth β-strand flanking the first strand.
Residues and secondary structures
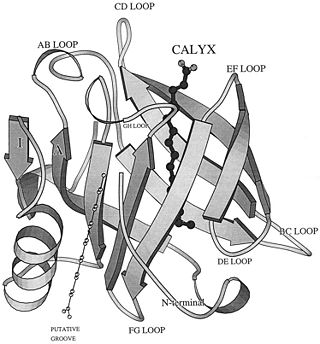
β-Lactoglobulin contains two disulfide bonds (Cys 66–Cys 160 and Cys 106–Cys 119) and a free thiol (Cys 121)(Fig. 1). Structures of βLG have been reported by several groups with X-ray crystallography and solution NMR, that it is predominantly β-sheet protein. The β-barrel, or so called calyx, is conical and is made of two β-sheets: the B–D strands and N-terminal half of the A strand (denoted AN) form one sheet, and the E–H strands and C-terminal half of the A strand (denoted AC) form the other. On the outer surface of the β-barrel, between the G and H strands, is the 3-turn α-helix. The loops that connect the β-strands at the closed end of the calyx, BC, DE, and FG, are generally quite short, whereas those at the open end, AB, CD, EF,and GH, are significantly longer and more flexible. In the calyx, there is a large central cavity which is surrounded by hydrophobic residues and is accessible to solvent. This cavity provides the principal ligand-binding site (Fig. 2). [10]
Genetic Variants
Genetically, β-lactoglobulin may exist as one of several variants, among which the variants A and B are the most abundant. The A and B variants of the protein differ from each other by amino acid residues at positions Asp64 (Gly64 in variant B) and Val118 (Ala118 in variant B). These differences in primary structure render the two variants slightly different with respect to isoelectric point, solubility, self-association properties, as well as pressure and temperature stability. However, the structural characteristics of the A and B variants of bovine b-lactoglobulin are virtually indistinguishable. In its native state, β-lactoglobulin is a predominantly β-sheet protein containing nine b-strands and three a-helices. The core of the protein is formed by a flattened b-barrel (a calyx) composed of eight antiparallel b-strands (A to H).[11]

Dimer/Monomer
At physiological conditions, majority of bovine b-lactoglobulin forms a dimer (Fig. 3). Below pH 3, the dimer dissociates into monomers which maintain their native conformation. Dimeric Lactoglobulin molecules exist in the open conformation at basic pH, whereas they exist in the closed conformation at acidic pH, after undergoing Tanford transition around neutral pH.[12]
Ligands and Active sites
The true function of β-Lg is unknown, but it has been suggested that it is involved in the transport of retinol and/or fatty acids. It binds retinol with a higher affinity than does RBP. β-Lg contains two tryptophans, Trp 19 on the A strand and Trp 61 on the C strand. The former is buried in the hydrophobic core whereas the latter is exposed to the solvent in the native structure, making them useful probes for monitoring site-specific conformational changes[13] and their fluorescence is altered when retinol is bound.[14] Co-crystallized β-Lg with palmitic acid (3D Model 2._1B0O) and the refined structure reveals that the ligand binds in the central cavity in a manner similar to the binding of retinol to the related lipocalin, serum retinol-binding protein. The carboxyl group binds to both Lys-60 and Lys-69 at the entrance to the cavity. The hydrophobic tail stretches in an almost fully extended conformation into the center of the protein.[15] In addition, studies on the monomer–dimer equilibrium and the reactivity of the thiol group of Cys121 which deeply buried between the α-helix and H strand revealed other important properties of β-LG. The stability of the structure also depend so heavily upon the external loop around residue 64 or the β strand with the free thiol.[16]

β-Lg has binding sites for hydrophobic ligands like fatty acids, retinoic acid, and Vitamin D3 (cholecalciferol) and lactose ligands [18] For hydrophobic ligands two sites have been postulated, one inside the calyx and the other at the dimer interface, on the outer surface of the protein between the α-helix and the β-barrel (Fig. 4). The accessibility to the calyx is pH-dependent. NMR and Xray analysis showed that the access is mediated by the mobile EF loop. All the structures with ligands bound to the calyx exhibit an open EF loop, suggesting that this site is accessible at neutral pH.
|
Biological function
Besides as binding protein, β-Lg is also assumed to have a transporter function especially for several ligands through the digestive tract in, as the calyx can protect any ligands entered/bound to the cavity.[19].
Molecular mechanism of the Tanford transition
The Tanford transition is a conformational change of bovine β-lactoglobulin occurring at around pH 7, identified originally on the basis of optical rotatory dispersion and the accessibility of a thiol group. X-ray analysis has suggested that a conformational change to the EF-loop (residues I84 -N90) (Fig. 5) is responsible for the Tanford transition, with the loop closing the hydrophobic cavity of the β-barrel of the β-LG molecule below pH 7 and flipping to open the cavity above pH 7.[20]Tanford transition is triggered by protonation of Glu89 exhibiting an anomalously high pKa value. The Tanford transition may involve some other structural changes as well. For example, the transition is accompanied by a change in the microenvironment of Tyr42 and causes an alteration in the relative orientation of monomers in the dimer by as much as 5 degrees, which breaks a number of intersubunit hydrogen bonds. [21]
The structures of the trigonal crystal form of bovine β-lactoglobulin variant A at pH 6.2, 7.1, and 8.2 have been determined by X-ray diffraction methods. The glutamate side chain of residue 89 is buried at pH 6.2 and becomes exposed at pH 7.1 and 8.2. This conformational change, involving the loop 84-90, provides a structural basis for a variety of pH-dependent chemical, physical, and spectroscopic phenomena, collectively known as the Tanford transition.[22]It was found that the stereochemical environment of Tyr42 changes significantly with pH variation between pH 7 and pH 8. This may provide a structural explanation for an as yet unexplained feature of the Tanford transition, namely the increase in exposure of a tyrosine residue.[23]
Uses / Application
Food Industry and Pharmacy
β-Lactoglobulin interaction with hydrophobic molecules and with other proteins, and its sensitivity to chemical, thermal and baric denaturation, all with a view to establishing relationships among structure, properties and functionality and its potential use in food and pharmacy industry.[24] Food Industry has been widely utilize β-LG to increase functionality (sensory, texture, nutritional) in commercial food and beverages application, and also as inexpensive source of protein to increase the overall protein content of their product. Pharmacy Industry is recently investigating and developing design of drugs that utilize β-LG as vehicle to transport antioxidants, drugs and vitamins to the gut.[25]
Antioxidant Nature
In the dairy industry, bovine milk is frequently heated for pasteurization (62.5°C for 30 min) and sterilization. This heating process may induce oxidative losses of proteins, unsaturated lipids, vitamins, active enzymes, and immunological factors. Cross-linking the free thiol groups of β-LG by heating (100 °C for 2 min), or chemically modifying the β-LG by carboxymethylation to block the thiol groups resulted in a substantial loss of antioxidant activity. The data suggest that Cys-121 plays an essential role in the antioxidant nature of β-LG. Because β-LG is extremely sensitive to thermal denaturation, to maintain its antioxidant nature, dairy products consumed daily should not be overheated in order to maintain its antioxidant nature.[26] Whey Protein whose majority constituent is β-Lactoglobulin, gives abundant source of branched chain amino acids especially Leucine which plays a key role in initiating transcription of protein synthesis that speed recovery and adaptation to stress (exercise) and also a free thiol Cys which is (postulated) utilized in the body to generate antioxidant glutathione.[27]
Other β-Lactoglobulin related 3D structures and complexes
2q2m - Bovine β-Lactoglobulin Native (Fig. 4)
1b8e - Crystal structure of the Bovine β-Lactoglobulin (Isoforms A and B) in orthorombic space group
1qg5 - Crystal structure of the Bovine β-Lactoglobulin (Isoforms A)
1beb - Bovine β-Lactoglobulin, Lattice X
1cj5 - Bovine β-Lactoglobulin A
1gx8 - Bovine β-Lactoglobulin complexed with Retinol, Trigonal Lattice Z
1gx9 - Bovine β-Lactoglobulin complexed with Retinoic acid, Trigonal Lattice Z
1gxa - Bovine β-Lactoglobulin complexed with Retinol and Palmitic acid, Trigonal Lattice Z
1b0o - Bovine β-Lactoglobulin complexed with Palmitate, Lattice Z
1bsy 2blg 3blg - Structural Basis of the Tanford Transitioon of Bovine β-Lactoglobulin from crystal structures at 3 pH values
References
- ↑ http://www.jbc.org/content/104/2/359.citation
- ↑ http://www.sciencedirect.com/science/article/pii/S0958694698000211
- ↑ Wu SY, Perez MD, Puyol P, Sawyer L. beta-lactoglobulin binds palmitate within its central cavity. J Biol Chem. 1999 Jan 1;274(1):170-4. PMID:9867826
- ↑ Kontopidis G, Holt C, Sawyer L. Invited review: beta-lactoglobulin: binding properties, structure, and function. J Dairy Sci. 2004 Apr;87(4):785-96. PMID:15259212 doi:http://dx.doi.org/10.3168/jds.S0022-0302(04)73222-1
- ↑ Flower DR, North AC, Sansom CE. The lipocalin protein family: structural and sequence overview. Biochim Biophys Acta. 2000 Oct 18;1482(1-2):9-24. PMID:11058743
- ↑ http://www.biochemj.org/bj/318/bj3180001.htm
- ↑ Newcomer ME, Jones TA, Aqvist J, Sundelin J, Eriksson U, Rask L, Peterson PA. The three-dimensional structure of retinol-binding protein. EMBO J. 1984 Jul;3(7):1451-4. PMID:6540172
- ↑ Liu HC, Chen WL, Mao SJ. Antioxidant nature of bovine milk beta-lactoglobulin. J Dairy Sci. 2007 Feb;90(2):547-55. PMID:17235131 doi:http://dx.doi.org/10.3168/jds.S0022-0302(07)71538-2
- ↑ Wu SY, Perez MD, Puyol P, Sawyer L. beta-lactoglobulin binds palmitate within its central cavity. J Biol Chem. 1999 Jan 1;274(1):170-4. PMID:9867826
- ↑ Sakurai K, Konuma T, Yagi M, Goto Y. Structural dynamics and folding of beta-lactoglobulin probed by heteronuclear NMR. Biochim Biophys Acta. 2009 Jun;1790(6):527-37. doi: 10.1016/j.bbagen.2009.04.003., Epub 2009 Apr 10. PMID:19362581 doi:http://dx.doi.org/10.1016/j.bbagen.2009.04.003
- ↑ Taulier N, Chalikian TV. Characterization of pH-induced transitions of beta-lactoglobulin: ultrasonic, densimetric, and spectroscopic studies. J Mol Biol. 2001 Dec 7;314(4):873-89. PMID:11734004 doi:http://dx.doi.org/10.1006/jmbi.2001.5188
- ↑ Vijayalakshmi L, Krishna R, Sankaranarayanan R, Vijayan M. An asymmetric dimer of beta-lactoglobulin in a low humidity crystal form-Structural changes that accompany partial dehydration and protein action. Proteins. 2007 Oct 11;71(1):241-249. PMID:17932936 doi:10.1002/prot.21695
- ↑ Sakurai K, Konuma T, Yagi M, Goto Y. Structural dynamics and folding of beta-lactoglobulin probed by heteronuclear NMR. Biochim Biophys Acta. 2009 Jun;1790(6):527-37. doi: 10.1016/j.bbagen.2009.04.003., Epub 2009 Apr 10. PMID:19362581 doi:http://dx.doi.org/10.1016/j.bbagen.2009.04.003
- ↑ Brownlow S, Morais Cabral JH, Cooper R, Flower DR, Yewdall SJ, Polikarpov I, North AC, Sawyer L. Bovine beta-lactoglobulin at 1.8 A resolution--still an enigmatic lipocalin. Structure. 1997 Apr 15;5(4):481-95. PMID:9115437
- ↑ Wu SY, Perez MD, Puyol P, Sawyer L. beta-lactoglobulin binds palmitate within its central cavity. J Biol Chem. 1999 Jan 1;274(1):170-4. PMID:9867826
- ↑ Brownlow S, Morais Cabral JH, Cooper R, Flower DR, Yewdall SJ, Polikarpov I, North AC, Sawyer L. Bovine beta-lactoglobulin at 1.8 A resolution--still an enigmatic lipocalin. Structure. 1997 Apr 15;5(4):481-95. PMID:9115437
- ↑ Dominguez-Ramirez L, Del Moral-Ramirez E, Cortes-Hernandez P, Garcia-Garibay M, Jimenez-Guzman J. beta-Lactoglobulin's Conformational Requirements for Ligand Binding at the Calyx and the Dimer Interphase: a Flexible Docking Study. PLoS One. 2013 Nov 8;8(11):e79530. doi: 10.1371/journal.pone.0079530. PMID:24255705 doi:http://dx.doi.org/10.1371/journal.pone.0079530
- ↑ Dominguez-Ramirez L, Del Moral-Ramirez E, Cortes-Hernandez P, Garcia-Garibay M, Jimenez-Guzman J. beta-Lactoglobulin's Conformational Requirements for Ligand Binding at the Calyx and the Dimer Interphase: a Flexible Docking Study. PLoS One. 2013 Nov 8;8(11):e79530. doi: 10.1371/journal.pone.0079530. PMID:24255705 doi:http://dx.doi.org/10.1371/journal.pone.0079530
- ↑ Kontopidis G, Holt C, Sawyer L. Invited review: beta-lactoglobulin: binding properties, structure, and function. J Dairy Sci. 2004 Apr;87(4):785-96. PMID:15259212 doi:http://dx.doi.org/10.3168/jds.S0022-0302(04)73222-1
- ↑ Sakurai K, Goto Y. Dynamics and mechanism of the Tanford transition of bovine beta-lactoglobulin studied using heteronuclear NMR spectroscopy. J Mol Biol. 2006 Feb 17;356(2):483-96. Epub 2005 Dec 1. PMID:16368109 doi:http://dx.doi.org/10.1016/j.jmb.2005.11.038
- ↑ Taulier N, Chalikian TV. Characterization of pH-induced transitions of beta-lactoglobulin: ultrasonic, densimetric, and spectroscopic studies. J Mol Biol. 2001 Dec 7;314(4):873-89. PMID:11734004 doi:http://dx.doi.org/10.1006/jmbi.2001.5188
- ↑ Qin BY, Bewley MC, Creamer LK, Baker HM, Baker EN, Jameson GB. Structural basis of the Tanford transition of bovine beta-lactoglobulin. Biochemistry. 1998 Oct 6;37(40):14014-23. PMID:9760236 doi:10.1021/bi981016t
- ↑ Oliveira KM, Valente-Mesquita VL, Botelho MM, Sawyer L, Ferreira ST, Polikarpov I. Crystal structures of bovine beta-lactoglobulin in the orthorhombic space group C222(1). Structural differences between genetic variants A and B and features of the Tanford transition. Eur J Biochem. 2001 Jan;268(2):477-83. PMID:11168385
- ↑ http://www.sciencedirect.com/science/article/pii/S0958694698000211
- ↑ http://www.faqs.org/patents/app/20110038942
- ↑ Liu HC, Chen WL, Mao SJ. Antioxidant nature of bovine milk beta-lactoglobulin. J Dairy Sci. 2007 Feb;90(2):547-55. PMID:17235131 doi:http://dx.doi.org/10.3168/jds.S0022-0302(07)71538-2
- ↑ http://www.kettlercise.com/members-area/supplements/whey-protein.html

