Nitrate reductase
From Proteopedia
(Difference between revisions)
| (17 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
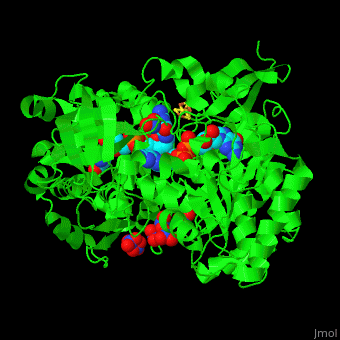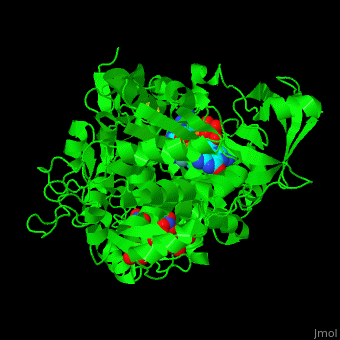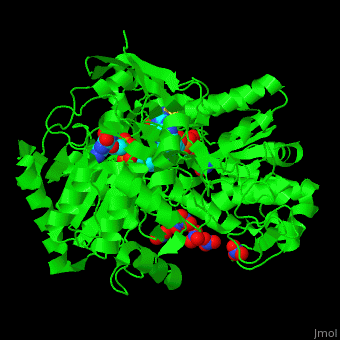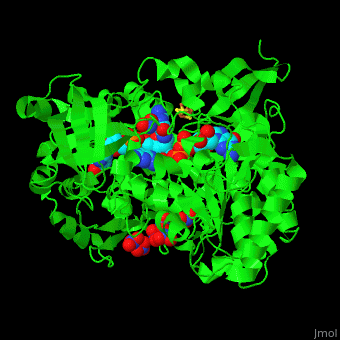
| - | + | <StructureSection load='' size='350' side='right' caption='Bacterial periplasmic dissimilatory nitrate reductase containing Fe4S4 cluster with Mo, NO3- and molybdepterin guanine nucleotide (PDB entry [[2jiq]])' scene='49/490062/Cv/3'> | |
| - | + | == Function == | |
| - | + | '''Nitrate reductase''' (NR) catalyzes the reduction of NO<sub>3</sub> to NO<sub>2</sub> using NADPH<ref>PMID:13152110</ref>. NR active site contains Mo atom. Four types of NR are known:<br /> | |
| - | + | '''eukaryotic assimilatory NR'''<br /> | |
| - | + | '''bacterial cytoplasmic assimilatory''' (NAC)<br /> | |
| - | + | '''bacterial membrane-bound respiratory''' (NAR)<br /> | |
| - | '''Nitrate reductase''' (NR) catalyzes the reduction of | + | '''bacterial periplasmic dissimilatory''' (NAP).<br /> |
| + | The cofactors of NR are Molybdopterin (MPT) in eukaryotic NR and bis-Molybdopterin guanine dinucleotide (MGD) in bacterial NR. | ||
| + | |||
| + | == Structural highlights == | ||
| + | <scene name='49/490062/Cv/7'>Bacterial periplasmic dissimilatory nitrate reductase containing Fe4S4 cluster with Mo, NO3 and 2 molybdepterin guanine nucleotides</scene>. NAP active site includes a <scene name='49/490062/Cv/8'>six-coordinated Mo atom which coordinates to 2 MGD molecules and Cys140</scene>. The <scene name='49/490062/Cv/9'>NO3 molecule is guided to the active site by a funnel leading to the Mo atom</scene><ref>PMID:18327621</ref>. | ||
| + | |||
| + | </StructureSection> | ||
==3D structures of nitrate reductase== | ==3D structures of nitrate reductase== | ||
| - | + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | |
| + | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| + | |||
| + | *Bacterial periplasmic dissimilatory NR | ||
| + | |||
| + | **[[2nap]], [[2jim]], [[2jio]], [[2jip]], [[2v3v]], [[2v45]] – DdNAP + MGD + Mo - ''Desulfovibrio desulfuricans''<br /> | ||
| + | **[[2jiq]] - DdNAP + NO3 + MGD + Mo<br /> | ||
| + | **[[2jir]] - DdNAP + CN + MGD + Mo<br /> | ||
| + | **[[1jni]] – NAP small subunit + heme – ''Haemophilus influenzae''<br /> | ||
| + | **[[1ogy]] - NAP + heme + MGD + Mo– ''Rhodobacter sphaeroides''<br /> | ||
| + | **[[2nya]] - EcNAP + MGD + Mo – ''Escherichia coli''<br /> | ||
| + | **[[3ml1]], [[3o5a]] - NAP catalytic subunit + NAPB + heme + MGD + Mo – ''Ralstonia eutropha''<br /> | ||
| + | **[[5yly]] – NR cytochrome b5 reductase domain + FAD – ''Ulva prolifera''<br /> | ||
| + | |||
| + | *Bacterial respiratory NR | ||
| - | [[ | + | **[[1q16]], [[1y5l]] – EcNAR α+β+γ chains + heme + MGD + Mo<br /> |
| - | [[ | + | **[[1r27]] - EcNAR α+β chains + MGD + Mo<br /> |
| - | [[ | + | **[[1siw]] – EcNAR α+β+γ chains + heme<br /> |
| - | [[ | + | **[[1y4z]], [[3egw]] - EcNAR α+β (mutant) + γ chains + inhibitor + heme + MGD + Mo<br /> |
| - | [[ | + | **[[1y5i]], [[1y5n]] - EcNAR α+β + γ (mutant) chains + inhibitor + heme + MGD + Mo<br /> |
| - | [[ | + | **[[3ir5]], [[3ir7]] - EcNAR α (mutant) +β + γ chains + inhibitor + heme + MGD + Mo<br /> |
| - | [[ | + | **[[3ir6]] - EcNAR α (mutant) +β + γ chains + inhibitor + heme + GDP |
| - | + | *Eukaryotic assimilatory NR | |
| - | [[ | + | **[[2bih]], [[2bii]] – NR MoCo-binding domain + MPT – ''Pichia angusta'' |
| - | [[ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | *Eukaryotic NR | |
| - | [[ | + | **[[1cne]] – cNR + FAD – corn<br /> |
| + | **[[1cnf]], [[2cnd]] – cNR (mutant) + FAD + ADP<br /> | ||
| + | }} | ||
| + | == References == | ||
| + | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
3D structures of nitrate reductase
Updated on 23-July-2019
References
- ↑ NICHOLAS DJ, NASON A. Molybdenum and nitrate reductase. II. Molybdenum as a constituent of nitrate reductase. J Biol Chem. 1954 Mar;207(1):353-60. PMID:13152110
- ↑ Najmudin S, Gonzalez PJ, Trincao J, Coelho C, Mukhopadhyay A, Cerqueira NM, Romao CC, Moura I, Moura JJ, Brondino CD, Romao MJ. Periplasmic nitrate reductase revisited: a sulfur atom completes the sixth coordination of the catalytic molybdenum. J Biol Inorg Chem. 2008 Mar 8;. PMID:18327621 doi:10.1007/s00775-008-0359-6