This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Alpha-1-antitrypsin
From Proteopedia
(Difference between revisions)
| (8 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
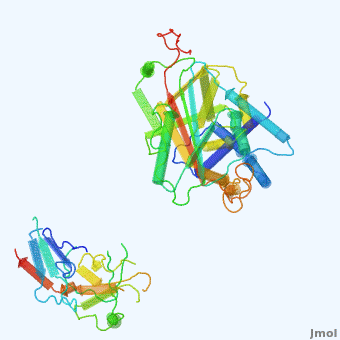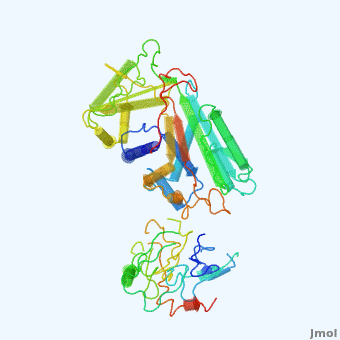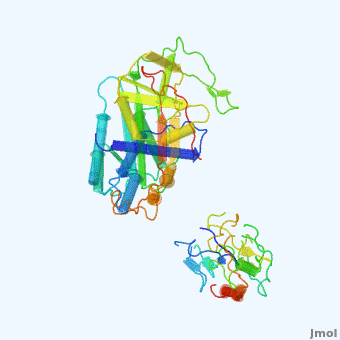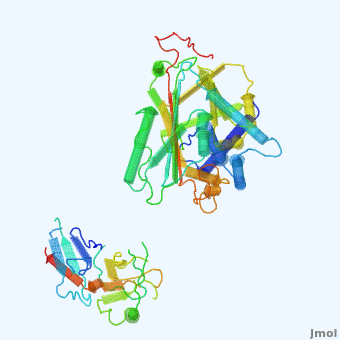
| - | <StructureSection load='' size=' | + | <StructureSection load='' size='350' side='right' scene='User:Daniel_Seeman/Alpha-1-antitrypsin/437437437437/1' caption='Transition of α1-antitrypsin between active and inactive conformations '> |
| + | __TOC__ | ||
| + | === Function === | ||
| + | |||
'''Alpha-1-antitrypsin''' (also known as α1-antitrypsin or A1AT) is an inhibitor of [[Elastase]] and [[Trypsin]]. It is a member of the '''Ser'''ine '''P'''rotease '''I'''nhibitor ([[:Category:Serpin|Serpin]]) family, and as such undergoes a conformational change where the substrate protein associates with a loop region on A1AT causing that loop to become ordered as a Beta Strand<ref name="nature_paper">''Nature'' '''455''', 1189-1190 (30 October 2008)</ref>. In this case Trypsin (the substrate) is inhibited when a covalent bond is formed to A1AT through the newly formed Beta region<ref name="nature_paper" />. Once bound covalently to its substrate the stability of the A1AT complex goes up drastically, making it an effective "molecular mousetrap"<ref name="nature_paper" />. With A1AT, as with most members of the Serpin family, the transition from inactive precursor protein to active complex comes after a cleavage event<ref name="nature_paper" />. | '''Alpha-1-antitrypsin''' (also known as α1-antitrypsin or A1AT) is an inhibitor of [[Elastase]] and [[Trypsin]]. It is a member of the '''Ser'''ine '''P'''rotease '''I'''nhibitor ([[:Category:Serpin|Serpin]]) family, and as such undergoes a conformational change where the substrate protein associates with a loop region on A1AT causing that loop to become ordered as a Beta Strand<ref name="nature_paper">''Nature'' '''455''', 1189-1190 (30 October 2008)</ref>. In this case Trypsin (the substrate) is inhibited when a covalent bond is formed to A1AT through the newly formed Beta region<ref name="nature_paper" />. Once bound covalently to its substrate the stability of the A1AT complex goes up drastically, making it an effective "molecular mousetrap"<ref name="nature_paper" />. With A1AT, as with most members of the Serpin family, the transition from inactive precursor protein to active complex comes after a cleavage event<ref name="nature_paper" />. | ||
Shown <scene name='User:Daniel_Seeman/Alpha-1-antitrypsin/437437437437/1'>on the right</scene> is a morph, generated by the <span class="plainlinks">[http://molmovdb.mbb.yale.edu/molmovdb/morph/ Yale Morph Server]</span> that shows A1AT going from its inactive form, to the conformation in which it is bound to Trypsin (also shown in the same animation)<ref>The <span class="plainlinks">[http://molmovdb.mbb.yale.edu/molmovdb/morph/ Yale Morph Server]</span></ref>. | Shown <scene name='User:Daniel_Seeman/Alpha-1-antitrypsin/437437437437/1'>on the right</scene> is a morph, generated by the <span class="plainlinks">[http://molmovdb.mbb.yale.edu/molmovdb/morph/ Yale Morph Server]</span> that shows A1AT going from its inactive form, to the conformation in which it is bound to Trypsin (also shown in the same animation)<ref>The <span class="plainlinks">[http://molmovdb.mbb.yale.edu/molmovdb/morph/ Yale Morph Server]</span></ref>. | ||
| Line 11: | Line 14: | ||
*<scene name='User:Daniel_Seeman/Alpha-1-antitrypsin/A1at_test/1'>A1AT as represented by pdb ID 1atu</scene> | *<scene name='User:Daniel_Seeman/Alpha-1-antitrypsin/A1at_test/1'>A1AT as represented by pdb ID 1atu</scene> | ||
*<scene name='User:Daniel_Seeman/Alpha-1-antitrypsin/A1at_test2/1'>A1AT bound to Trypsin (its substrate)</scene> | *<scene name='User:Daniel_Seeman/Alpha-1-antitrypsin/A1at_test2/1'>A1AT bound to Trypsin (its substrate)</scene> | ||
| - | </StructureSection> | ||
| - | __NOTOC__ | ||
==3D structures of Alpha-1-antitrypsin== | ==3D structures of Alpha-1-antitrypsin== | ||
| + | [[Alpha-1-antitrypsin 3D structures]] | ||
| - | + | </StructureSection> | |
| - | + | __NOTOC__ | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Nature 455, 1189-1190 (30 October 2008)
- ↑ The Yale Morph Server
- ↑ Biochemistry, Fifth Edition, p.289.
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Daniel Seeman, Alexander Berchansky, Joel L. Sussman