Sandbox Reserved 819
From Proteopedia
(Difference between revisions)
| (93 intermediate revisions not shown.) | |||
| Line 3: | Line 3: | ||
<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | <!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
| - | =''' | + | |
| - | + | ||
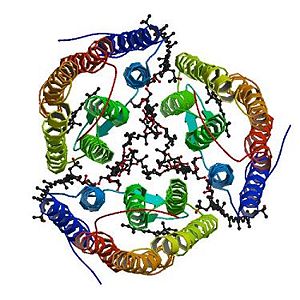
| + | <StructureSection load='2z55' size='300' side='right' caption='2z55: cristal made of four Archaerhodopsin-2' scene=''> | ||
| + | =2Z55= | ||
__TOC__ | __TOC__ | ||
| + | ==Introduction== | ||
| - | ==I kept a copy of your first version, tell me if you prefer it== | ||
| - | + | Archaerhodopsin-2 (aR2) is a light-driven proton pump. The resulting proton gradient is subsequently converted into chemical energy. | |
| + | The structure in 3D represented here is 2Z55, a cristal made of four archaerhodopsin-2. | ||
| + | Archaerhodopsin-2 is composed of 259 amino acids. 88% of this amino acid sequence is identical to the sequence of the archaerhodopsin-1. Moreover, there is 56% identity between this sequence and the sequence of the bacteriorhodopsin. <ref name="seq">PMID: 1654776</ref> | ||
| + | ==The trimeric structure of Archaerhodopsin-2== | ||
| - | + | [[Image:2z55_bio_r_500.jpg|300px|left|thumb|The trimeric structure with its ligands]] | |
| + | Archaerhodopsin-2 is a retinal protein–carotenoid complex found in the claret membrane of Halorubrum sp. aus-2 and it represents a real adaptation to life at high salt concentrations. In these membranes, three Archaerhodopsin-2 chains form a trimeric structure [http://www.pdb.org/pdb/explore/jmol.do?structureId=2Z55&view=symmetry&bionumber=1] (the image on the left side represents the trimeric structure), capturing light energy and using it to move protons across the membrane out of the cell. It exists four different chains with different structures: A,B,D,E (they are not represented here). | ||
| + | The trimerization increases the thermal stability of the protein aR2 in the claret membrane of Halorubrum sp. aus-2 and enlarges the pH range where the protein can keep its neutral conformation. Thus, a larger pH gradient can be generated across the membrane, leading to an increased efficiency of the proton pumping. Therefore the trimeric structure is more efficient than the monomeric structure. | ||
| - | The trimeric structure functions as a light-driven proton pump thanks to | + | Archaerhodopsin-2 consists of the protein moiety rhodopsin and a reversibly covalently bound cofactor, the retinal. |
| + | The trimeric structure functions as a light-driven proton pump thanks to this retinal molecule, called <scene name='56/568017/New_scene_ret_2/1'>RET</scene>, which changes its conformation when absorbing a photon, resulting in a conformational change of the surrounding protein and the proton pumping action. | ||
| - | + | Others ligands are linked with each subunit of the trimeric structure like the bacterioruberin (<scene name='56/568017/22b/1'>22B</scene>). The bacterioruberin plays a structural role for the trimerization of aR2. | |
| + | Several saccharides, some lipids and glycolipids also interact with the trimeric structure like the 2,3-di-phytanyl-glycerol (<scene name='56/568017/New_scene_3/1'>L2P</scene>). The lipids and the glycolipids fill the intratrimer hydrophobic space and they are required to the complex activity. Others lipids surround the trimeric structure and are essential to preserve it.<ref name="multiple">PMID:18082767</ref> | ||
| + | |||
| + | ===The rhodopsin=== | ||
| + | |||
| + | |||
| + | The rhodopsin belongs to the CATH Superfamily 1.20.1070.10.[http://www.cathdb.info/version/3.5.0/superfamily/1.20.1070.10]The protein <scene name='56/568017/Rhodopsin/1'>rhodopsin</scene> has 7 transmembrane alpha helices, embedded in the plasma membrane. These helices are connected to each other by protein loops. | ||
| - | + | The rhodopsin harvests energy from light to carry out metabolic processes using a non-chlorophyll-based pathway. Thanks to the retinal, the light induces a phototactic response by interacting with transducer membrane-embedded proteins that have no relation to G proteins. There are four different rhodopsins with different structures: A, B, D, E. | |
| Line 29: | Line 43: | ||
| - | The | + | The retinal (C20 H28 O) is a photoreactive chromophore. |
| - | + | The rhodopsin binds retinal [http://www.ebi.ac.uk/pdbe-srv/pdbechem/chemicalCompound/show/RET] in a central pocket on the seventh helix by a covalent bond with the <scene name='56/568017/Lysine_221/1'>lysine residue 221</scene>. Others bonds exist, like van-der-waals bonds [http://www.ebi.ac.uk/pdbe-site/pdbemotif/?tab=boundmolecule&pdb=2z55&ligandCode3letter=RET]. | |
| + | Retinal is a polyene chromophore and allows to convert light into metabolic energy. It absorbs visible light maximally at 550-570 nm. | ||
| + | It catches a photon, leading to a conformational change of the rhodopsin. This is due to an isomerization of the 11-cis-retinal into a 11-trans-retinal. The retinal binds covalently to the lysine 221 on the transmembrane helix nearest the C-terminus of the protein through a Schiff base linkage. Formation of the Schiff base linkage involves removing the oxygen atom from retinal and two hydrogen atoms from the free amino group of lysine, giving H2O. Retinylidene is the divalent group formed by removing the oxygen atom from retinal, and so opsins is called retinylidene proteins. A Schiff base is a compound with a functional group made up of a carbon-nitrogen double bond with a nitrogen atom connected to an aryl or alkyl group, not hydrogen. Schiff bases in a broad sense have the general formula R1-R2-C=N-R3, where R is an organic side chain. In this definition, Schiff base is synonymous with azomethine. The chain on the nitrogen makes the Schiff base a stable imine. A Schiff base is derived from an aniline, where R3 is a phenyl or a substituted phenyl. {{Wikipedia|Schiff_base}} | ||
| + | |||
| - | == | + | ==Ligands== |
| - | The rhodopsin belongs to the CATH Superfamily 1.20.1070.10[http://www.cathdb.info/version/3.5.0/superfamily/1.20.1070.10] | ||
| + | ===The bacterioruberin (22B)=== | ||
| - | The rhodopsin harvests energy from light to carry out metabolic processes using a non-chlorophyll-based pathway.The light induces a phototactic response, thanks to the retinal, by interacting with transducer membrane-embedded proteins that have no relation to G proteins. There are four different rhodopsin with different structures: A, B, D, E. | ||
| + | The bacterioruberin [http://www.ebi.ac.uk/pdbe-srv/pdbechem/chemicalCompound/show/22B] (C50 H76 O4) is a 50 carbon carotenoid pigment which give a red color to the membrane. The primary role of bacterioruberin in the cell is to protect against DNA damage incurred by UV light.<ref>Shammohammadi, H.R., Protective roles of bacterioruberin and intracellular KCl in the resistance of ''Halobacterium salinarium'' against DNA-damaging agents, J Radiat Res, 1998, 39(4):251.</ref> This protection is not, however, due to the ability of bacterioruberin to absorb UV light. Bacterioruberin protects the DNA by acting as an antioxidant, rather than directly blocking UV light.<ref>Ide, H., Takeshi, S., Hiroaki, T., Studies on the antioxidation activity of bacterioruberin, Urakami Found Mem, 1998, 6:127–33.</ref>It is able to protect the cell from reactive oxygen species produced from exposure to UV by acting as a target. | ||
| + | |||
| + | Furthermore, the bacterioruberin is essential because it plays a structural role for the trimerization of aR2: it mediates interactions between neighbouring monomers. | ||
| + | When the bacterioruberin is bound to the Archaerhodopsin-2, its polyene chain is between the A and B helices of one monomere and the D and E helices of an adjacent one. One end of the bacterioruberin is next to the cytoplasmic membrane surfaces and thus is able to interact with the hydrophilic residus of two monomers. The other end of the bacterioruberin protrudes out of the extracellular membrane. <ref name="multiple">PMID:18082767</ref> | ||
| + | More precisely, it binds to: the B chain thanks to a hydrogen bond with the <scene name='56/568017/Threonine_112/1'>Threonine 112</scene>, the <scene name='56/568017/Tyrosine/1'>Tyrosine 156</scene> and thanks to an electrostatic bond with the HOH 304; the D chain thanks to a hydrogen bond with the Tyrosine 156; the E chain thanks to a hydrogen bond with the Tyrosine 156. Others bonds exist like van-der-waals bonds [http://www.ebi.ac.uk/pdbe-site/pdbemotif/?tab=boundmolecule&pdb=2z55&ligandCode3letter=22B]. | ||
| - | ==Ligands== | ||
| - | ===The bacterioruberin (22B)=== | ||
| + | ===Glycolipids=== | ||
| - | + | ||
| - | + | Some glycolipids seem to bind to the trimeric structure. They are in the centre of the three monomers and the hydrophilic part seems to be composed of three hexoses connected in tandems. | |
| - | + | These glycolipids are supposed to have a stabilizing effect on the trimeric structure because each head group interacts with the two neighbouring monomers. <ref name="multiple">PMID:18082767</ref> | |
| Line 54: | Line 74: | ||
| - | The 2,3-di-phytanyl-glycerol [http://www.ebi.ac.uk/pdbe-srv/pdbechem/chemicalCompound/show/L2P] | + | The 2,3-di-phytanyl-glycerol [http://www.ebi.ac.uk/pdbe-srv/pdbechem/chemicalCompound/show/L2P] (C43 H88 O3) is an archaeol (di-O-phytanylglycerol). This is a double ether of sn-1-glycerol where positions 2 and 3 are bound to phytanyl residues. The archaeols are Archaea homologs of diacylglycerols (DAGs). |
| - | It interacts with the aR2 surface and the carbohydrate <scene name='56/568017/Glc/1'>GLC</scene>. It binds to: the A chain thanks a covalent bond with the carbohydrate alpha-D-glucose 281 (GLC) and thanks a hydrogen bond with the <scene name='56/568017/ | + | |
| + | It interacts with the aR2 surface and the carbohydrate <scene name='56/568017/Glc/1'>GLC</scene>. It binds to: the A chain thanks to a covalent bond with the carbohydrate alpha-D-glucose 281 (GLC) and thanks to a hydrogen bond with the <scene name='56/568017/Tyr85/1'>Tyrosine 85</scene>; the B chain thanks to a covalent bond with the carbohydrate alpha-D-glucose 281 (GLC) and thanks to a hydrogen bond with the Tyrosine 85; the D chain thanks to a covalent bond with the carbohydrate alpha-D-glucose 281 (GLC) and thanks to a hydrogen bond with the Tyrosine 85; the E chain thanks to a covalent bond with the carbohydrate alpha-D-glucose 284 (GLC) and thanks to a hydrogen bond with the Tyrosine 85. Others bonds exist like van-der-waals bonds [http://www.ebi.ac.uk/pdbe-site/pdbemotif/?tab=boundmolecule&pdb=2z55&ligandCode3letter=L2P]. | ||
| + | |||
===Saccharides=== | ===Saccharides=== | ||
| - | == | + | |
| + | |||
| + | Several saccharides can interact with the trimeric structure: β-D-galactose (<scene name='56/568017/Gal/1'>GAL</scene>) [http://www.ebi.ac.uk/pdbe-srv/pdbechem/chemicalCompound/show/GAL], α-D-glucose (<scene name='56/568017/Glc/1'>GLC</scene>) [http://www.ebi.ac.uk/pdbe-srv/pdbechem/chemicalCompound/show/GLC] and α-D-mannose (<scene name='56/568017/Man/1'>MAN</scene>) [http://www.ebi.ac.uk/pdbe-srv/pdbechem/chemicalCompound/show/MAN]. | ||
| + | |||
| + | A molecule of α-D-mannose is covalently bound to one α-D-glucose and one β-D-galactose,thus creating a <scene name='56/568017/Glc-man-gal_and_rhodopsin/1'>glucose-mannose-galactose (GLC-MAN-GAL) ligand</scene>. There is one ligand by monomer. The glucose can create a covalent bond with the molecule of 2,3-Di-Phytanyl-Glycerol. | ||
| + | |||
| + | |||
| + | ==Comparison between the Archaerhodopsin-2 and the Bacteriorhodopsin== | ||
| + | |||
| + | |||
| + | 56% of the Archaeorhodopsin-2 sequence is identical to the [[Bacteriorhodopsin]] sequence.<ref name="seq">PMID: 1654776</ref> | ||
| + | |||
| + | Most amino acids that play a role in the trimerization are not conserved between the two proteins. For instance, the counterparts of some hydrophobic residues of the Archaerhodopsin-2 (the one interacting with the polyene chain of the bacterioruberin) have a different volume. Another difference is the fact that the polar residues of the Archaerhodopsin-2 (responsible for the hydrogen bonds with the bacterioruberin) are replaced in the Bacteriorhodopsin by hydrophobic amino acids. | ||
| + | |||
| + | However the global structures of Archaeorhodopsin-2 and Bacteriorhodopsin are really similar, especially at the level of the open space between the monomers. The interaction between the monomers of the Bacteriorhodopsin is also mediated by lipids: diphytanyl diether phospholipids instead of Bacterioruberin. | ||
| + | |||
| + | This similarity of structure forms the basis of several hypothesis concerning the mechanisms of the Archaeorhodopsin-2 <ref name="multiple">PMID:18082767</ref> | ||
| + | </StructureSection> | ||
| + | |||
| + | == 3D structures of Archaerhodopsin-2 and Bacteriorhodopsin == | ||
| + | |||
| + | |||
| + | [[2ei4]]-Trimeric structure of Archaerhodopsin-2 | ||
| + | |||
| + | [[1vgo]]-Crystal Structure of Archaerhodopsin-2 | ||
| + | |||
| + | [[1uaz]]-Crystal structure of archaerhodopsin-1 | ||
| + | |||
| + | [[1iw6]]-Crystal Structure of the Ground State of Bacteriorhodopsin | ||
| + | |||
== References == | == References == | ||
<references /> | <references /> | ||
| + | |||
| + | |||
| + | == Proteopedia page contributors and editors == | ||
| + | |||
| + | Lydwine Germain, Allan Bernard | ||
Current revision
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
| |||||||||||
3D structures of Archaerhodopsin-2 and Bacteriorhodopsin
2ei4-Trimeric structure of Archaerhodopsin-2
1vgo-Crystal Structure of Archaerhodopsin-2
1uaz-Crystal structure of archaerhodopsin-1
1iw6-Crystal Structure of the Ground State of Bacteriorhodopsin
References
- ↑ 1.0 1.1 Uegaki K, Sugiyama Y, Mukohata Y. Archaerhodopsin-2, from Halobacterium sp. aus-2 further reveals essential amino acid residues for light-driven proton pumps. Arch Biochem Biophys. 1991 Apr;286(1):107-10. PMID:1654776
- ↑ 2.0 2.1 2.2 2.3 Yoshimura K, Kouyama T. Structural role of bacterioruberin in the trimeric structure of archaerhodopsin-2. J Mol Biol. 2008 Feb 1;375(5):1267-81. Epub 2007 Nov 22. PMID:18082767 doi:10.1016/j.jmb.2007.11.039
- ↑ Shammohammadi, H.R., Protective roles of bacterioruberin and intracellular KCl in the resistance of Halobacterium salinarium against DNA-damaging agents, J Radiat Res, 1998, 39(4):251.
- ↑ Ide, H., Takeshi, S., Hiroaki, T., Studies on the antioxidation activity of bacterioruberin, Urakami Found Mem, 1998, 6:127–33.
Proteopedia page contributors and editors
Lydwine Germain, Allan Bernard