A-ATP Synthase
From Proteopedia
(Difference between revisions)
| (One intermediate revision not shown.) | |||
| Line 1: | Line 1: | ||
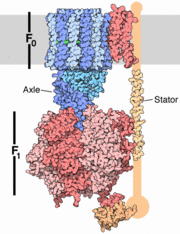
| - | <StructureSection load=1e79 size='450' side='right' caption='Bovine ATP synthase chains α (grey, green, pink), β (yellow, magenta, cyan), γ (gold), δ (red), ε (wheat) (PDB code [[1e79]])' scene=''> | + | <StructureSection load=1e79 size='450' side='right' caption='Bovine ATP synthase chains α (grey, green, pink), β (yellow, magenta, cyan), γ (gold), δ (red), ε (wheat) complex with ATP (stick model), ADP (stick model), glycerol, dicyclohexylurea, sulfate and Mg+2 ions (green) (PDB code [[1e79]])' scene=''> |
[[Image:F_ATPsynthase.gif | thumb|left]] | [[Image:F_ATPsynthase.gif | thumb|left]] | ||
__NOTOC__ | __NOTOC__ | ||
| Line 34: | Line 34: | ||
Residue <scene name='A-ATP_Synthase/238/6'>S238</scene> is a polar serine molecule that interacts with the nucleotides via a hydrogen bond during catalysis. The distance between residue S238 is longest in '''As''', shortest in '''Avi''' and intermediate in '''Apnp''' . In '''As''' a water molecule bridges the gap, which is removed in '''Avi'''. Dehydration of the transition state active site is reversed when ATP forms. In '''Apnp''' the water molecule interacts with the y-phosphate of ATP. | Residue <scene name='A-ATP_Synthase/238/6'>S238</scene> is a polar serine molecule that interacts with the nucleotides via a hydrogen bond during catalysis. The distance between residue S238 is longest in '''As''', shortest in '''Avi''' and intermediate in '''Apnp''' . In '''As''' a water molecule bridges the gap, which is removed in '''Avi'''. Dehydration of the transition state active site is reversed when ATP forms. In '''Apnp''' the water molecule interacts with the y-phosphate of ATP. | ||
| - | == | + | ==Comparisons to other known structures== |
This P-loop has an arched conformation unique to A-ATP Synthase, indicating that the mode of nucleotide binding and the catalytic mechanism is different from that of other syntheses. <ref name= Priya> PMID: 21925186</ref> | This P-loop has an arched conformation unique to A-ATP Synthase, indicating that the mode of nucleotide binding and the catalytic mechanism is different from that of other syntheses. <ref name= Priya> PMID: 21925186</ref> | ||
Current revision
| |||||||||||
References
- ↑ 1.0 1.1 Muller V, Lemker T, Lingl A, Weidner C, Coskun U, Gruber G. Bioenergetics of archaea: ATP synthesis under harsh environmental conditions. J Mol Microbiol Biotechnol. 2005;10(2-4):167-80. PMID:16645313 doi:10.1159/000091563
- ↑ Gibbons C, Montgomery MG, Leslie AG, Walker JE. The structure of the central stalk in bovine F(1)-ATPase at 2.4 A resolution. Nat Struct Biol. 2000 Nov;7(11):1055-61. PMID:11062563 doi:10.1038/80981
- ↑ Schafer IB, Bailer SM, Duser MG, Borsch M, Bernal RA, Stock D, Gruber G. Crystal structure of the archaeal A1Ao ATP synthase subunit B from Methanosarcina mazei Go1: Implications of nucleotide-binding differences in the major A1Ao subunits A and B. J Mol Biol. 2006 May 5;358(3):725-40. Epub 2006 Mar 10. PMID:16563431 doi:http://dx.doi.org/10.1016/j.jmb.2006.02.057
- ↑ Gonzalez JM, Masuchi Y, Robb FT, Ammerman JW, Maeder DL, Yanagibayashi M, Tamaoka J, Kato C. Pyrococcus horikoshii sp. nov., a hyperthermophilic archaeon isolated from a hydrothermal vent at the Okinawa Trough. Extremophiles. 1998 May;2(2):123-30. PMID:9672687
- ↑ 5.0 5.1 Manimekalai MS, Kumar A, Jeyakanthan J, Gruber G. The Transition-Like State and P(i) Entrance into the Catalytic A Subunit of the Biological Engine A-ATP Synthase. J Mol Biol. 2011 Mar 16. PMID:21396943 doi:10.1016/j.jmb.2011.03.010
- ↑ Priya R, Kumar A, Manimekalai MS, Gruber G. Conserved Glycine Residues in the P-Loop of ATP Synthases Form a Doorframe for Nucleotide Entrance. J Mol Biol. 2011 Sep 8. PMID:21925186 doi:10.1016/j.jmb.2011.08.045
Proteopedia Page Contributors and Editors (what is this?)
Kaitlin Chase MacCulloch, Michal Harel, Alexander Berchansky