Sandbox Reserved 819
From Proteopedia
(Difference between revisions)
| (9 intermediate revisions not shown.) | |||
| Line 2: | Line 2: | ||
{{Sandbox_Reserved_ESBS}} | {{Sandbox_Reserved_ESBS}} | ||
<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | <!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
| + | |||
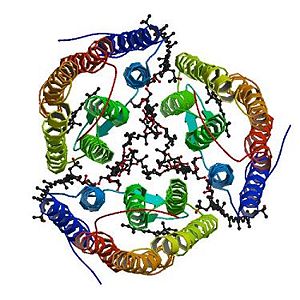
<StructureSection load='2z55' size='300' side='right' caption='2z55: cristal made of four Archaerhodopsin-2' scene=''> | <StructureSection load='2z55' size='300' side='right' caption='2z55: cristal made of four Archaerhodopsin-2' scene=''> | ||
| + | =2Z55= | ||
| + | __TOC__ | ||
==Introduction== | ==Introduction== | ||
| Line 73: | Line 76: | ||
The 2,3-di-phytanyl-glycerol [http://www.ebi.ac.uk/pdbe-srv/pdbechem/chemicalCompound/show/L2P] (C43 H88 O3) is an archaeol (di-O-phytanylglycerol). This is a double ether of sn-1-glycerol where positions 2 and 3 are bound to phytanyl residues. The archaeols are Archaea homologs of diacylglycerols (DAGs). | The 2,3-di-phytanyl-glycerol [http://www.ebi.ac.uk/pdbe-srv/pdbechem/chemicalCompound/show/L2P] (C43 H88 O3) is an archaeol (di-O-phytanylglycerol). This is a double ether of sn-1-glycerol where positions 2 and 3 are bound to phytanyl residues. The archaeols are Archaea homologs of diacylglycerols (DAGs). | ||
| - | It interacts with the aR2 surface and the carbohydrate <scene name='56/568017/Glc/1'>GLC</scene>. It binds to: the A chain thanks to a covalent bond with the carbohydrate alpha-D-glucose 281 (GLC) and thanks to a hydrogen bond with the <scene name='56/568017/ | + | It interacts with the aR2 surface and the carbohydrate <scene name='56/568017/Glc/1'>GLC</scene>. It binds to: the A chain thanks to a covalent bond with the carbohydrate alpha-D-glucose 281 (GLC) and thanks to a hydrogen bond with the <scene name='56/568017/Tyr85/1'>Tyrosine 85</scene>; the B chain thanks to a covalent bond with the carbohydrate alpha-D-glucose 281 (GLC) and thanks to a hydrogen bond with the Tyrosine 85; the D chain thanks to a covalent bond with the carbohydrate alpha-D-glucose 281 (GLC) and thanks to a hydrogen bond with the Tyrosine 85; the E chain thanks to a covalent bond with the carbohydrate alpha-D-glucose 284 (GLC) and thanks to a hydrogen bond with the Tyrosine 85. Others bonds exist like van-der-waals bonds [http://www.ebi.ac.uk/pdbe-site/pdbemotif/?tab=boundmolecule&pdb=2z55&ligandCode3letter=L2P]. |
| Line 89: | Line 92: | ||
56% of the Archaeorhodopsin-2 sequence is identical to the [[Bacteriorhodopsin]] sequence.<ref name="seq">PMID: 1654776</ref> | 56% of the Archaeorhodopsin-2 sequence is identical to the [[Bacteriorhodopsin]] sequence.<ref name="seq">PMID: 1654776</ref> | ||
| - | Most amino acids that play a role in the trimerization are not conserved between the two proteins. For instance, the counterparts of some hydrophobic residues of the Archaerhodopsin-2 (the one interacting with the polyene chain of the bacterioruberin) have a different volume. Another difference is the fact that the | + | Most amino acids that play a role in the trimerization are not conserved between the two proteins. For instance, the counterparts of some hydrophobic residues of the Archaerhodopsin-2 (the one interacting with the polyene chain of the bacterioruberin) have a different volume. Another difference is the fact that the polar residues of the Archaerhodopsin-2 (responsible for the hydrogen bonds with the bacterioruberin) are replaced in the Bacteriorhodopsin by hydrophobic amino acids. |
| - | However the global structures of Archaeorhodopsin-2 and | + | However the global structures of Archaeorhodopsin-2 and Bacteriorhodopsin are really similar, especially at the level of the open space between the monomers. The interaction between the monomers of the Bacteriorhodopsin is also mediated by lipids: diphytanyl diether phospholipids instead of Bacterioruberin. |
This similarity of structure forms the basis of several hypothesis concerning the mechanisms of the Archaeorhodopsin-2 <ref name="multiple">PMID:18082767</ref> | This similarity of structure forms the basis of several hypothesis concerning the mechanisms of the Archaeorhodopsin-2 <ref name="multiple">PMID:18082767</ref> | ||
Current revision
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
| |||||||||||
3D structures of Archaerhodopsin-2 and Bacteriorhodopsin
2ei4-Trimeric structure of Archaerhodopsin-2
1vgo-Crystal Structure of Archaerhodopsin-2
1uaz-Crystal structure of archaerhodopsin-1
1iw6-Crystal Structure of the Ground State of Bacteriorhodopsin
References
- ↑ 1.0 1.1 Uegaki K, Sugiyama Y, Mukohata Y. Archaerhodopsin-2, from Halobacterium sp. aus-2 further reveals essential amino acid residues for light-driven proton pumps. Arch Biochem Biophys. 1991 Apr;286(1):107-10. PMID:1654776
- ↑ 2.0 2.1 2.2 2.3 Yoshimura K, Kouyama T. Structural role of bacterioruberin in the trimeric structure of archaerhodopsin-2. J Mol Biol. 2008 Feb 1;375(5):1267-81. Epub 2007 Nov 22. PMID:18082767 doi:10.1016/j.jmb.2007.11.039
- ↑ Shammohammadi, H.R., Protective roles of bacterioruberin and intracellular KCl in the resistance of Halobacterium salinarium against DNA-damaging agents, J Radiat Res, 1998, 39(4):251.
- ↑ Ide, H., Takeshi, S., Hiroaki, T., Studies on the antioxidation activity of bacterioruberin, Urakami Found Mem, 1998, 6:127–33.
Proteopedia page contributors and editors
Lydwine Germain, Allan Bernard