Sandbox Reserved 813
From Proteopedia
(Difference between revisions)
| (3 intermediate revisions not shown.) | |||
| Line 2: | Line 2: | ||
{{Sandbox_Reserved_ESBS}} | {{Sandbox_Reserved_ESBS}} | ||
<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | <!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
| + | <big>'''OCT-4 or POU5F1'''</big><StructureSection load='3l1p' size='500' side='right' caption='Structure of Oct-4 (PDB entry [[3l1p]])' scene=''> | ||
| - | |||
| - | <big>'''OCT-4 or POU5F1'''</big> | ||
| - | {{STRUCTURE_3l1p| PDB=3l1p | SCENE= }} | ||
= Introduction = | = Introduction = | ||
[http://en.wikipedia.org/wiki/Oct-4 Oct-4] for Octamer–binding transcription factor 4, also known as POU5F1 ('''POU domain, class 5, transcription factor 1''') is a 352 amino acid protein encoded by the ''POU5F1'' gene. It is the earliest expressed gene known, and the gene is developmentally regulated during mammalian embryogenesis. The map position of oct 4 on mouse chromosome 17 is between Q and T region in the Major histocompatibility. The Oct-4 POU transcription factor is expressed in mammalian totipotent embryonic stem and germ cells. Oct-4 is the only factor that cannot be replace by an other factor of the POU family to induce pluripotency. His remplacement by a transcription factor cocktail does not lead to the successful derivation of induced pluripotent stem cells. | [http://en.wikipedia.org/wiki/Oct-4 Oct-4] for Octamer–binding transcription factor 4, also known as POU5F1 ('''POU domain, class 5, transcription factor 1''') is a 352 amino acid protein encoded by the ''POU5F1'' gene. It is the earliest expressed gene known, and the gene is developmentally regulated during mammalian embryogenesis. The map position of oct 4 on mouse chromosome 17 is between Q and T region in the Major histocompatibility. The Oct-4 POU transcription factor is expressed in mammalian totipotent embryonic stem and germ cells. Oct-4 is the only factor that cannot be replace by an other factor of the POU family to induce pluripotency. His remplacement by a transcription factor cocktail does not lead to the successful derivation of induced pluripotent stem cells. | ||
| Line 49: | Line 47: | ||
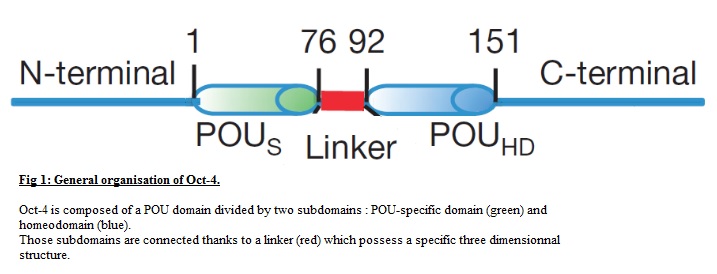
== Linker== | == Linker== | ||
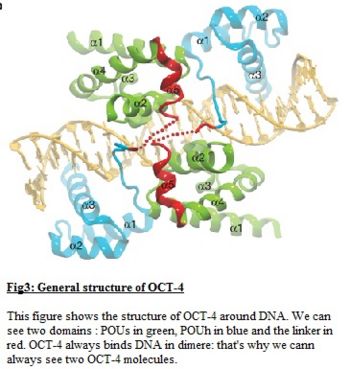
| - | <scene name='56/568011/Linker/1'>The linker</scene> tethers the two POU subdomains and is hypervariable in both sequence and length. Thus the linker in Oct-4 contrary to the linker of the other member of the POU family like Oct-1 [http://en.wikipedia.org/wiki/POU2F1], is a 17 amino acid alpha-helix and is exposed to the protein surface. The | + | <scene name='56/568011/Linker/1'>The linker</scene> tethers the two POU subdomains and is hypervariable in both sequence and length. Thus the linker in Oct-4 contrary to the linker of the other member of the POU family like Oct-1 [http://en.wikipedia.org/wiki/POU2F1], is a 17 amino acid alpha-helix and is exposed to the protein surface. The Oct-4 linker functions as a '''protein-protein interaction''' interface and plays a highly important role during '''reprogramming'''. This alpha 5 helix interacts with helices alpha 2 and alpha 4 of POUs mostly by Van der Waals interactions, except for the hydrogen bond between<scene name='56/568011/Liaison_25-81/1'>Tyr 25 of POUs and Gln 81</scene> of the linker. The interaction between Val 36 of POUs and the the carboxy-terminal of the helix alpha 5 of the linker plays an important role. In all known Oct-4 sequences <scene name='56/568011/Asn79_and_leu80/1'>Asn79 and Leu80 </scene> are invariant but not conserved in other members of Oct family. Mutation in the linker of GLn81 by an Arg leads to a complete loss-of-function phenotype. Likewise mutation of Leu80 by an Ala abolished biological activity. It is deduced that this mutation disturbs an interaction surface with yet unknown additional factors. Overall mutation of the linker of the protein lead <scene name='56/568011/Asn76_asn_77_asn_79/1'>residues exposed to the surface</scene>(Asn76, Asn 77, Asn 79) to significantly fewer function. The integrity of the linker is essential for successful reprogramming. |
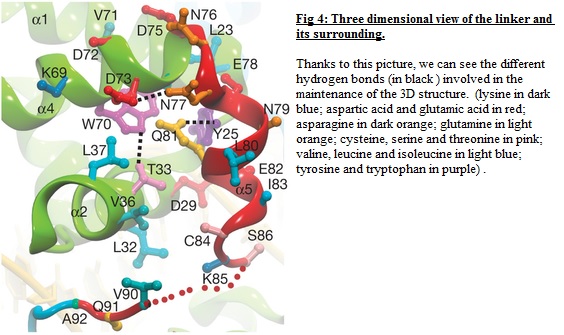
On the figure n°4, we can see the surrounding of the linker. There are different hydrogen bonds to save the tridimentional structure of OCT-4: the first hydrogen bond is located between <scene name='56/568011/Hydrogen_bond_between_33-_70/1'> Tyr 33 and the Try70 </scene>. | On the figure n°4, we can see the surrounding of the linker. There are different hydrogen bonds to save the tridimentional structure of OCT-4: the first hydrogen bond is located between <scene name='56/568011/Hydrogen_bond_between_33-_70/1'> Tyr 33 and the Try70 </scene>. | ||
| Line 59: | Line 57: | ||
==Pou-homeodomain== | ==Pou-homeodomain== | ||
| - | The <scene name='56/568011/Pouhd/1'>homeodomain</scene> (POUhd) is a very conserved 60 amino acid '''three helices alpha DNA-binding domain'''. We can find the homeodomain | + | The <scene name='56/568011/Pouhd/1'>homeodomain</scene> (POUhd) is a very conserved 60 amino acid '''three helices alpha DNA-binding domain'''. We can find the homeodomain from the 223th to the 282th amino acid in the Oct-4 sequence. Its DNA sequence is called “homeobox”, and the genes are known as “Hox gene”. The DNA recognition helix (alpha 3) binds the DNA major groove while the amino-terminal tail binds the DNA minor groove. We can draw a parallel with oct1 and oct2 about the Homeodomain and admit that the ninth residue of the DNA-recognition helix is a cysteine that might have been conserved at this position to confer DNA binding specificity , and promote more relaxed DNA sequence recognition by POU domains. The recognition helix and the inter-helix loops are rich in arginine and lysine residues, which form hydrogen bonds with the DNA backbone. |
| Line 65: | Line 63: | ||
==In vivo== | ==In vivo== | ||
| - | Oct-4 is able to create a complex with Sox-2 to express genes involved in the embryonic development such as YES1. Oct-4 is also involved in the creation of intestinal and skin cancer. Indeed, the overexpression of Oct-4 provokes the upregulation of B-catenin [http://en.wikipedia.org/wiki/Beta-catenin] transcription which inhibit the cellular differentiation. | + | Oct-4 is able to create a complex with Sox-2 to express genes involved in the embryonic development such as ''YES1''. Oct-4 is also involved in the creation of intestinal and skin cancer. Indeed, the overexpression of Oct-4 provokes the upregulation of B-catenin [http://en.wikipedia.org/wiki/Beta-catenin] transcription which inhibit the cellular differentiation. |
==In vitro== | ==In vitro== | ||
| - | Oct-4, Sox2, [[c-Myc]] and Klf4 are the four factors involved in the '''cell reprogamming'''. Indeed, those four factors were used in the Yamanaka’s studies to reprogram mature cells in '''inducted pluripotent stem cells''' ([http://fr.wikipedia.org/wiki/IPS iPS] )and are thus also named “Yamanaka factors”. When they are in a mature cell, they can induce the expression of genes involved in the reprogramation of cells in an embryonic-state: cells which possess the same morphology and the same growth properties than embryonic stem cells. The unique structure of the linker domain between the POU-specific domain and the Pou-homeodomain gives Oct-4 a protein very important for the study of iPS cells . Indeed, the others transcription factors are recruited and a complex is created with the linker domain. | + | Oct-4, Sox2, [[c-Myc]] and Klf4 are the four factors involved in the '''cell reprogamming'''. Indeed, those four factors were used in the Yamanaka’s studies to reprogram mature cells in '''inducted pluripotent stem cells''' ([http://fr.wikipedia.org/wiki/IPS iPS] )and are thus also named “Yamanaka factors”. When they are in a mature cell, they can induce the expression of genes involved in the reprogramation of cells in an embryonic-state: cells which possess the same morphology and the same growth properties than embryonic stem cells. The unique structure of the linker domain between the POU-specific domain and the Pou-homeodomain gives Oct-4 a protein very important for the study of iPS cells .Indeed, the others transcription factors are recruited and a complex is created with the linker domain. |
= References = | = References = | ||
Current revision
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
| |||||||||||