Context
SRP54 is a 54kDa cytosolic protein part of the Signal Recognition Particle (SRP).
SRP54 bacterial homologous can be referred to as Ffh (Fifty Four Homologous).
SRP is a ribonucleoprotein particle essential for the translation and integration in membranes of signal peptide-bearing proteins. It is composed of an RNA backbone, on which bind different proteins.
For example Mammalian SRP are formed of the RNA 7S and SRP 9-14-19-54-68-72.
SRP54 and regions of RNA 7S are highly conserved in every organism and it was found that these two components are sufficient to form a minimal SRP.
The role of SRP is to :
- Bind signal peptide bearing proteins at the beginning of their translation by the ribosome (at this step we call the ribosome and the protein it began to translate the Ribosome Nascent Chain – RNC)
- Stop protein elongation
- Dock the RNC to a SRP Receptor (SR) and insert the protein into a translocon channel. This final step is GTP-dependant.
SRP54 is responsible for the signal peptide binding and the GTP-dependant interaction with SR. This protein is therefore essential in the SRP mechanism resulting in the integration of proteins into membranes or their secretion.
SRP54 is a 504 aminoacids protein,composed of 3 domains:
- N-terminal domain with 4 alpha-helices;
- G domain, central, with a GTPase activity
- M domain ( for Methionin Rich).
The 1QB2 structure is the M-domain of the human SRP54. It includes the aminoacids 322 to 441.
The following parts will treat of 1QB2 structure and its relation to the functions of SRP54M.
Structure
SRP54M is a 120 aminoacids long polypeptide. 1QB2 is a , since the studied polypeptide has the interesting property to dimerize in solution.
Note : for a readability purpose the structures enlighten in the Jmol applet will focus only on one SRP54. The same structures are present in the second SRP54M of the dimer.
The secondary structure of human SRP54M is formed of . The helices 2 to 7 form the , stabilized by hydrophobic, hydrogen and ionic interactions.
Several residues important to maintain the Core structure were identified. Among them the
Met382 is invariable while Glu386, Arg402 and Arg405 are well-conserved but not systemically found in the SRP54M of different organisms.
The remaining helix, , is not part of the Core and protrudes from it.
Between the helices are loops of various importance.
The loop including the , besides having an important role in SRP54 function, has the particularity to have two phenylalanine residues, , stacking their aromatic cycles. The function of this loop will be developed in the next part.
Fonction
The signal peptide binds SRP54M in an formed by helices 2, 3 and 4 as well as the loop 349-365 (17 aminoacids) previously mentioned.
These structures circumscribe a deep elongated hydrophobic groove matching the shape and chemical properties of known signal peptides.
Interestingly enough, the dimer structure of 1QB2 shows that the
The study of the H1 helix indicates that it shares common physical and chemical properties with alpha-helical signal peptides.
Theses similarities might come from the fact that the H1 helix is near the hydrophobic groove of its own SRP54M, and that conformational changes (in a full SRP54 protein) might bring H1 into the groove in order to protect it from solvent interaction in the absence of signal peptide.
It was therefore hypothesized that the binding of H1 into the groove of another SRP54M provides a possible model of the interaction between the signal peptide and SRP54M in vivo.
It also explains the dimerization of SRP54M in solution.
This hypothesis has however to be confirmed since 1QB2 is only a fragment of SRP54, and that the M-domain, linked to the N and G domains, might adopt another conformation.
SRP54M binds the SRP RNA 7S by electrostatic interactions.
The residues responsible for this interaction are localized in , but most particularly in helices 5 and 6.
A large part of the top of SRP54M is therefore positively charged to bind with the negatively charged 7S RNA.
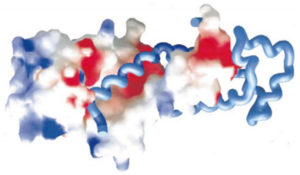
The helices 4, 5, 6 and 7 surfaces are colored in blue on the spacefill model (left) of SRP54M in this picture. For more details see figure 7 of reference 1.
It was also shown that the SRP54M - SRP RNA interaction is highly dependent of the structural integrity of the .
Experiments conducted with shorter versions of the M-domain, lacking some aminoacids of the Core, showed a loss in SRP RNA binding activity.
Despite being charged positively and close to the regions interacting with SRP RNA, highly conserved do not seem to interact with the negatively charged RNA.
This is explained by the fact that theses residues and their lateral chains are inside the core, and can not interact directly with the RNA. They are however important for the conformation of the core.
Related Structures
1e8o, 1e8s - hSRP9 + hSRP14 + 7S RNA – human
1ry1 - hSRP9 + hSRP14 + hSRP19 + hSRP54 + 7S RNA
1jid – hSRP19 + RNA
3ktv - hSRP19 + RNA S domain
1mfq – hSRP19 + hSRP54 M domain + 7S RNA S domain
References
- Clemons, W M, Jr, K Gowda, S D Black, C Zwieb, and V Ramakrishnan. “Crystal Structure of the Conserved Subdomain of Human Protein SRP54M at 2.1 A Resolution: Evidence for the Mechanism of Signal Peptide Binding.” Journal of Molecular Biology 292, no. 3 (September 24, 1999): 697–705. doi:10.1006/jmbi.1999.3090.
- Halic, Mario, Thomas Becker, Martin R. Pool, Christian M. T. Spahn, Robert A. Grassucci, Joachim Frank, and Roland Beckmann. “Structure of the Signal Recognition Particle Interacting with the Elongation-Arrested Ribosome.” Nature 427, no. 6977 (February 26, 2004): 808–814. doi:10.1038/nature02342.
- Huang, Qiaojia, Sayran Abdulrahman, Jiaming Yin, and Christian Zwieb. “Systematic Site-Directed Mutagenesis of Human Protein SRP54: Interactions with Signal Recognition Particle RNA and Modes of Signal Peptide Recognition.” Biochemistry 41, no. 38 (September 24, 2002): 11362–11371.
Contributors
Gregoire de Claviere and Hamelin Baptiste

