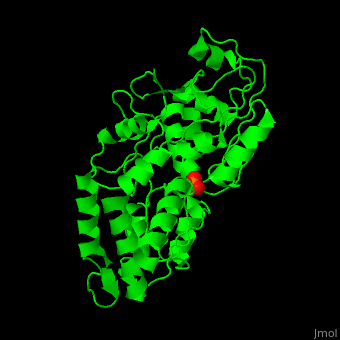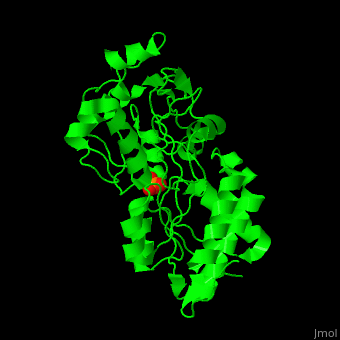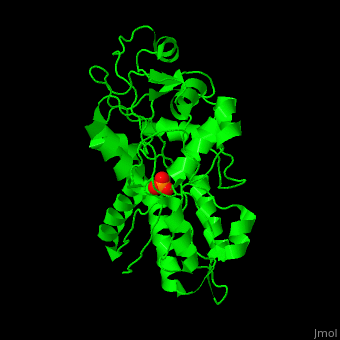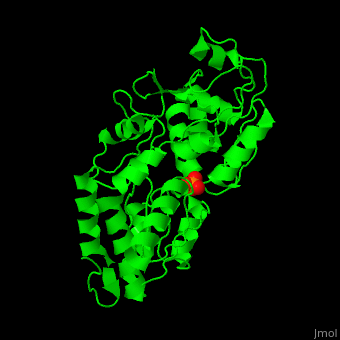
Phosphate-binding protein
From Proteopedia
| (10 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='' size='350' side='right' scene='48/488458/Cv/1' caption='E. coli phosphate-binding protein complex with phosphate [[1pbp]]'> | |
| - | + | == Function == | |
| - | + | '''Phosphate-binding protein''' (PBP) binds phosphate (Pi) with high affinity. PBP is involved in various biological processes like cell cycle regulation. PBP is produced under conditions of low Pi concentration. It binds Pi in the periplasm and transfers it to a membrane protein which transports it to the cytoplasm. PBP undergoes conformational change upon binding to Pi<ref>PMID:25338617</ref>. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | '''Phosphate-binding protein''' (PBP) binds phosphate (Pi) with high affinity. PBP is involved in various biological processes like cell cycle regulation. PBP is produced under conditions of low Pi concentration. It binds Pi in the periplasm and transfers it to a membrane protein which transports it to the cytoplasm. PBP undergoes conformational change upon binding to Pi. | + | |
| + | == Structural highlights == | ||
| + | The <scene name='48/488458/Cv/4'>PBP phosphate-binding site</scene> is highly selective. It can bind monobasic and dibasic phosphate but does not bind sulfate<ref>PMID:7929197</ref>. | ||
| + | </StructureSection> | ||
==3D structures of phosphate-binding protein== | ==3D structures of phosphate-binding protein== | ||
Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| + | [[2v3q]] – hPBP + Pi – human<br /> | ||
[[1qui]], [[1oib]] - EcPBP (mutant) - ''Escherichia coli''<br /> | [[1qui]], [[1oib]] - EcPBP (mutant) - ''Escherichia coli''<br /> | ||
| - | [[4jwo]] – PBP – ''Planctomyces limnophilus''<br /> | ||
| - | [[4exl]], [[4h1x]] – SpPBP – ''Streptococcus pneumoniae''<br /> | ||
[[1pbp]], [[2abh]], [[1ixh]] – EcPBP + Pi <br /> | [[1pbp]], [[2abh]], [[1ixh]] – EcPBP + Pi <br /> | ||
[[1qui]] - EcPBP (mutant) + Br + Pi<br /> | [[1qui]] - EcPBP (mutant) + Br + Pi<br /> | ||
[[1quj]], [[1qul]] - EcPBP (mutant) + Cl + Pi<br /> | [[1quj]], [[1qul]] - EcPBP (mutant) + Cl + Pi<br /> | ||
| - | [[1quk]], [[1ixi]], [[1ixg]], [[ | + | [[1quk]], [[1ixi]], [[1ixg]], [[1a40]] - EcPBP (mutant) + Pi<br /> |
[[1a54]], [[1a55]] - EcPBP (mutant) + dihydrogenphosphate<br /> | [[1a54]], [[1a55]] - EcPBP (mutant) + dihydrogenphosphate<br /> | ||
| - | [[2v3q]] – hPBP + Pi – human<br /> | ||
[[1pc3]], [[4lvq]] – PBP + Pi – ''Mycobacterium tuberculosis''<br /> | [[1pc3]], [[4lvq]] – PBP + Pi – ''Mycobacterium tuberculosis''<br /> | ||
[[3w9v]], [[3w9w]] – upPBP + Pi – unidentified prokaryote<br /> | [[3w9v]], [[3w9w]] – upPBP + Pi – unidentified prokaryote<br /> | ||
| + | [[4m1v]] - upPBP (mutant) + Pi<br /> | ||
| + | [[4exl]], [[4h1x]] – SpPBP – ''Streptococcus pneumoniae''<br /> | ||
[[4lat]] – SpPBP + Pi <br /> | [[4lat]] – SpPBP + Pi <br /> | ||
| - | [[ | + | [[4omb]], [[4pqj]] – PBP + Pi – ''Pseudomonas aeruginosa''<br /> |
| + | [[5wnn]] – PBP + Pi – ''Brucella melitensis''<br /> | ||
| + | [[5ub3]] – XcPBP – ''Xanthomonas citri''<br /> | ||
| + | [[5i84]], [[5ub4]] – XcPBP + Pi <br /> | ||
| + | [[5ub6]] – XcPBP + PPi <br /> | ||
| + | [[5ub7]] – XcPBP + ATP<br /> | ||
| + | [[5j1d]] – PBP + Pi – ''Stenotrophomonas maltophilia''<br /> | ||
| + | [[4jwo]] – PBP – ''Planctomyces limnophilus''<br /> | ||
| + | == References == | ||
| + | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
3D structures of phosphate-binding protein
Updated on 22-September-2020
2v3q – hPBP + Pi – human
1qui, 1oib - EcPBP (mutant) - Escherichia coli
1pbp, 2abh, 1ixh – EcPBP + Pi
1qui - EcPBP (mutant) + Br + Pi
1quj, 1qul - EcPBP (mutant) + Cl + Pi
1quk, 1ixi, 1ixg, 1a40 - EcPBP (mutant) + Pi
1a54, 1a55 - EcPBP (mutant) + dihydrogenphosphate
1pc3, 4lvq – PBP + Pi – Mycobacterium tuberculosis
3w9v, 3w9w – upPBP + Pi – unidentified prokaryote
4m1v - upPBP (mutant) + Pi
4exl, 4h1x – SpPBP – Streptococcus pneumoniae
4lat – SpPBP + Pi
4omb, 4pqj – PBP + Pi – Pseudomonas aeruginosa
5wnn – PBP + Pi – Brucella melitensis
5ub3 – XcPBP – Xanthomonas citri
5i84, 5ub4 – XcPBP + Pi
5ub6 – XcPBP + PPi
5ub7 – XcPBP + ATP
5j1d – PBP + Pi – Stenotrophomonas maltophilia
4jwo – PBP – Planctomyces limnophilus
References
- ↑ Gonzalez D, Richez M, Bergonzi C, Chabriere E, Elias M. Crystal structure of the phosphate-binding protein (PBP-1) of an ABC-type phosphate transporter from Clostridium perfringens. Sci Rep. 2014 Oct 16;4:6636. doi: 10.1038/srep06636. PMID:25338617 doi:http://dx.doi.org/10.1038/srep06636
- ↑ Wang Z, Choudhary A, Ledvina PS, Quiocho FA. Fine tuning the specificity of the periplasmic phosphate transport receptor. Site-directed mutagenesis, ligand binding, and crystallographic studies. J Biol Chem. 1994 Oct 7;269(40):25091-4. PMID:7929197