Palmitoyl protein thioesterase
From Proteopedia
(Difference between revisions)
| (7 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| + | {{BAMBED | ||
| + | |DATE=May 3, 2014 | ||
| + | |OLDID=1923505 | ||
| + | |BAMBEDDOI=10.1002/bmb.20840 | ||
| + | }} | ||
<StructureSection load='3GRO' size='450' side='right' caption='Human Palmitoyl-protein thioesterase 1 (PPT-1) homodimer (PDB: [[3gro]])' scene='43/436866/Overall-3-rainbow/1' > | <StructureSection load='3GRO' size='450' side='right' caption='Human Palmitoyl-protein thioesterase 1 (PPT-1) homodimer (PDB: [[3gro]])' scene='43/436866/Overall-3-rainbow/1' > | ||
| Line 4: | Line 9: | ||
'''<scene name='43/436866/Overall-3-rainbow/1'>Palmitoyl-protein thioesterase 1 (PPT-1)</scene>''' is a small glycoprotein [[hydrolase]] found in the lysosome that breaks the thioester bond between cysteine [[amino acids]] and <scene name='43/436866/Palmitic_acid_self/2'>palmitic acid</scene><ref name="newest">PMID:24083319</ref>. PPT-1 is a homodimer that is composed primarily of alpha helices and beta sheets with a <scene name='58/580839/Narrow_binding_groove/1'>hydrophobic groove</scene> that allows the [http://en.wikipedia.org/wiki/Palmitic_acid palmitic acid] to bind, exposing the thioester bond to the catalytic triad. PPT-1 was first found as an enzyme that removed palmitate from [[GTPase HRas]] and now has many additional cellular substrates <ref name="mutations">PMID:10781062</ref>. When PPT-1 is not functioning properly, lipid modified proteins can build up in the cells, causing lysosomal storage diseases and aiding in tumor formation <ref name="INCL">PMID:19302939</ref>. | '''<scene name='43/436866/Overall-3-rainbow/1'>Palmitoyl-protein thioesterase 1 (PPT-1)</scene>''' is a small glycoprotein [[hydrolase]] found in the lysosome that breaks the thioester bond between cysteine [[amino acids]] and <scene name='43/436866/Palmitic_acid_self/2'>palmitic acid</scene><ref name="newest">PMID:24083319</ref>. PPT-1 is a homodimer that is composed primarily of alpha helices and beta sheets with a <scene name='58/580839/Narrow_binding_groove/1'>hydrophobic groove</scene> that allows the [http://en.wikipedia.org/wiki/Palmitic_acid palmitic acid] to bind, exposing the thioester bond to the catalytic triad. PPT-1 was first found as an enzyme that removed palmitate from [[GTPase HRas]] and now has many additional cellular substrates <ref name="mutations">PMID:10781062</ref>. When PPT-1 is not functioning properly, lipid modified proteins can build up in the cells, causing lysosomal storage diseases and aiding in tumor formation <ref name="INCL">PMID:19302939</ref>. | ||
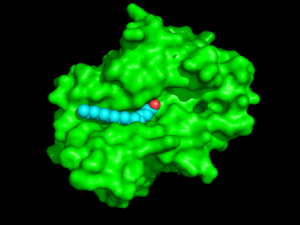
[[Image:Protopedia_surface_w_acid.png |300px|left|thumb|Figure 1: Surface view of PPT-1 (green) showing the hydrophobic groove and palmitate (blue with red oxygen)]] | [[Image:Protopedia_surface_w_acid.png |300px|left|thumb|Figure 1: Surface view of PPT-1 (green) showing the hydrophobic groove and palmitate (blue with red oxygen)]] | ||
| - | + | {{Clear}} | |
== Structure == | == Structure == | ||
The secondary structure of PPT1 contains several α-helices and few β-sheets (Figure 1). PPT1 includes residues 28-306, after the 27-residue signal peptide has been removed <ref name="RSCB">PMID:10781062</ref>. An insertion is found between β6 and β7, residues 140-223, and that forms a <scene name='57/573128/9/1'>second domain</scene>, shown in blue, that is compromised almost entirely of the fatty acid binding site. This second domain region contains six helices, α2-α7<ref name="RSCB"/>. | The secondary structure of PPT1 contains several α-helices and few β-sheets (Figure 1). PPT1 includes residues 28-306, after the 27-residue signal peptide has been removed <ref name="RSCB">PMID:10781062</ref>. An insertion is found between β6 and β7, residues 140-223, and that forms a <scene name='57/573128/9/1'>second domain</scene>, shown in blue, that is compromised almost entirely of the fatty acid binding site. This second domain region contains six helices, α2-α7<ref name="RSCB"/>. | ||
| Line 11: | Line 16: | ||
===Hydrophobic Groove === | ===Hydrophobic Groove === | ||
| - | The <scene name='57/573128/3/1'>hydrophobic binding groove</scene> is located in the second domain of PPT1 | + | The <scene name='57/573128/3/1'>hydrophobic binding groove</scene> where palmitate binds is located in the second domain of PPT1. (<scene name='48/489312/Binding_groove/4'>This alternate view</scene> shows the protein-ligand complex in the same orientation as Figure 1 above for better comparison). The fact that palmitate has to <scene name='57/573128/6/1'>bend</scene> to fit into the binding pocket suggests that this pocket is designed to bind an unsaturated fatty acid, with a possible cis-double bond between C4 and C5 (Figure 1)<ref name="RSCB"/>. The top portion of the groove is formed by the residues from α2 to α3. The acid binds in a [https://en.wikipedia.org/wiki/Gauche_effect gauche conformation] creating a <scene name='58/580839/Kink_in_acid/1'>kink </scene> in the acid chain. This bending suggests that PPT-1 was originally designed to react with an unsaturated fatty acid with cis-double bonds. Several residues that are present near the active site create the rest of the groove, including <scene name='57/573128/10/1'>Ile235, Val236, Gln116, Gly40, and Met41</scene><ref name="RSCB"/>. |
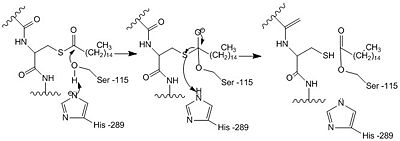
==Catalytic Triad== | ==Catalytic Triad== | ||
| Line 58: | Line 63: | ||
===Mutations leading to Juvenile Neuronal Ceroid Lipofuscinosis=== | ===Mutations leading to Juvenile Neuronal Ceroid Lipofuscinosis=== | ||
JNCL is caused by a mutation in the CLN3 gene which codes for a lysosomal membrane protein of unknown function <ref name="Ryan-3">PMID:9151311</ref>. Unlike the mutations that cause INCL and LINCL, mutations that lead to JNCL are located away from the active site and are seen to cause less damage to the overall structure of PPT-1. Some of the mutations in JNCL have been noted as retaining a low level of PPT-1 activity as the catalytic site is left fairly unperturbed. Mutations associated with JNCl are found in two locations, Thr75Pro with Asp79Gly and <scene name='58/580837/Juvenile_mutation/4'>Tyr247His with Gly250Val</scene>. These mutations occur in conjugate with its other pair and are predicted to disturb the geometry of helix α1, increasing the flexibility of the region, and alter the antiparallel βsheet motif in sheets βa and βb compared to the <scene name='58/580837/Tyrosine_normal/2'>normal Tyr-247 and Gly-250</scene>. | JNCL is caused by a mutation in the CLN3 gene which codes for a lysosomal membrane protein of unknown function <ref name="Ryan-3">PMID:9151311</ref>. Unlike the mutations that cause INCL and LINCL, mutations that lead to JNCL are located away from the active site and are seen to cause less damage to the overall structure of PPT-1. Some of the mutations in JNCL have been noted as retaining a low level of PPT-1 activity as the catalytic site is left fairly unperturbed. Mutations associated with JNCl are found in two locations, Thr75Pro with Asp79Gly and <scene name='58/580837/Juvenile_mutation/4'>Tyr247His with Gly250Val</scene>. These mutations occur in conjugate with its other pair and are predicted to disturb the geometry of helix α1, increasing the flexibility of the region, and alter the antiparallel βsheet motif in sheets βa and βb compared to the <scene name='58/580837/Tyrosine_normal/2'>normal Tyr-247 and Gly-250</scene>. | ||
| + | =3D structures of palmitoyl protein thioesterase= | ||
| + | [[Thioesterase]] | ||
</StructureSection> | </StructureSection> | ||
| Line 104: | Line 111: | ||
Audrey Wright | Audrey Wright | ||
| + | |||
| + | [[Category:Featured in BAMBED]] | ||
Current revision
This page, as it appeared on May 3, 2014, was featured in this article in the journal Biochemistry and Molecular Biology Education.
| |||||||||||
References
- ↑ Wang R, Borazjani A, Matthews AT, Mangum LC, Edelmann MJ, Ross MK. Identification of palmitoyl protein thioesterase 1 in human THP1 monocytes and macrophages and characterization of unique biochemical activities for this enzyme. Biochemistry. 2013 Oct 29;52(43):7559-74. doi: 10.1021/bi401138s. Epub 2013 Oct, 18. PMID:24083319 doi:http://dx.doi.org/10.1021/bi401138s
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 Bellizzi JJ 3rd, Widom J, Kemp C, Lu JY, Das AK, Hofmann SL, Clardy J. The crystal structure of palmitoyl protein thioesterase 1 and the molecular basis of infantile neuronal ceroid lipofuscinosis. Proc Natl Acad Sci U S A. 2000 Apr 25;97(9):4573-8. PMID:10781062 doi:10.1073/pnas.080508097
- ↑ 3.0 3.1 Simonati A, Tessa A, Bernardina BD, Biancheri R, Veneselli E, Tozzi G, Bonsignore M, Grosso S, Piemonte F, Santorelli FM. Variant late infantile neuronal ceroid lipofuscinosis because of CLN1 mutations. Pediatr Neurol. 2009 Apr;40(4):271-6. doi: 10.1016/j.pediatrneurol.2008.10.018. PMID:19302939 doi:http://dx.doi.org/10.1016/j.pediatrneurol.2008.10.018
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 Bellizzi JJ 3rd, Widom J, Kemp C, Lu JY, Das AK, Hofmann SL, Clardy J. The crystal structure of palmitoyl protein thioesterase 1 and the molecular basis of infantile neuronal ceroid lipofuscinosis. Proc Natl Acad Sci U S A. 2000 Apr 25;97(9):4573-8. PMID:10781062 doi:10.1073/pnas.080508097
- ↑ 5.0 5.1 5.2 Branneby, Cecilia. "Exploiting Enzyme Promiscuity for Rational Design." KTH Biotechnology (2005): Web. 10 Apr. 2013.
- ↑ 6.0 6.1 Hofmann SL, Das AK, Yi W, Lu JY, Wisniewski KE. Genotype-phenotype correlations in neuronal ceroid lipofuscinosis due to palmitoyl-protein thioesterase deficiency. Mol Genet Metab. 1999 Apr;66(4):234-9. PMID:10191107 doi:http://dx.doi.org/10.1006/mgme.1999.2803
- ↑ 7.0 7.1 Dawson G, Schroeder C, Dawson PE. Palmitoyl:protein thioesterase (PPT1) inhibitors can act as pharmacological chaperones in infantile Batten disease. Biochem Biophys Res Commun. 2010 Apr 23;395(1):66-9. doi:, 10.1016/j.bbrc.2010.03.137. Epub 2010 Mar 25. PMID:20346914 doi:http://dx.doi.org/10.1016/j.bbrc.2010.03.137
- ↑ Vines DJ, Warburton MJ. Classical late infantile neuronal ceroid lipofuscinosis fibroblasts are deficient in lysosomal tripeptidyl peptidase I. FEBS Lett. 1999 Jan 25;443(2):131-5. PMID:9989590
- ↑ Mitchison HM, Taschner PE, Kremmidiotis G, Callen DF, Doggett NA, Lerner TJ, Janes RB, Wallace BA, Munroe PB, O'Rawe AM, Gardiner RM, Mole SE. Structure of the CLN3 gene and predicted structure, location and function of CLN3 protein. Neuropediatrics. 1997 Feb;28(1):12-4. PMID:9151311 doi:http://dx.doi.org/10.1055/s-2007-973656
Similar Proteopedia Pages
Category:Palmitoyl_protein_thioesterase
External Resources
Gauche Effect Wikipedia page
Palmitic acid Wikipedia page
Infantile Neuronal Ceroid Lipofuscinosis Wikipedia page
PMSF Wikipedia page
Protein Chaperones Wikipedia page
Endoplasmic reticulum Wikipedia page
Palmitoylation Wikipedia page
Page on Late Infantile neuronal Ceroid Lipofuscinosis
Page on Juvenile neuronal Ceroid Lipofuscinosis
Student Contributors
Andrew Bartels
Nicole Green
Kelly Maddalone
Ryan Mughmaw
Audrey Wright
Proteopedia Page Contributors and Editors (what is this?)
R. Jeremy Johnson, Karsten Theis, Alexander Berchansky, Angel Herraez, Michal Harel