We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
3m9s
From Proteopedia
(Difference between revisions)
| (5 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| + | |||
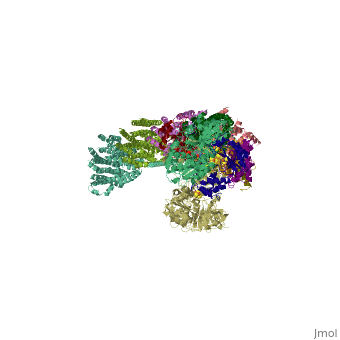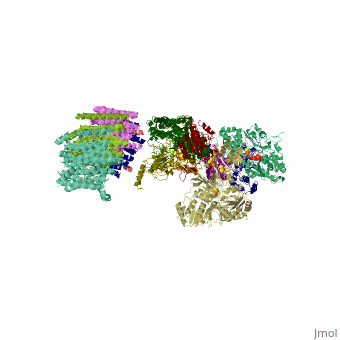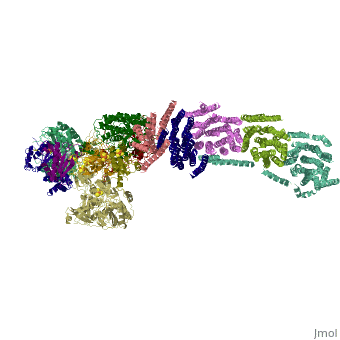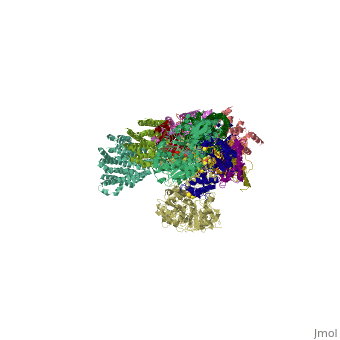
==Crystal structure of respiratory complex I from Thermus thermophilus== | ==Crystal structure of respiratory complex I from Thermus thermophilus== | ||
| - | <StructureSection load='3m9s' size='340' side='right' caption='[[3m9s]], [[Resolution|resolution]] 4.50Å' scene=''> | + | <StructureSection load='3m9s' size='340' side='right'caption='[[3m9s]], [[Resolution|resolution]] 4.50Å' scene=''> |
== Structural highlights == | == Structural highlights == | ||
| - | <table><tr><td colspan='2'>[[3m9s]] is a | + | <table><tr><td colspan='2'>[[3m9s]] is a 20 chain structure with sequence from [https://en.wikipedia.org/wiki/Thermus_thermophilus_HB8 Thermus thermophilus HB8]. The December 2011 RCSB PDB [https://pdb.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/index.html Molecule of the Month] feature on ''Complex I'' by David Goodsell is [https://dx.doi.org/10.2210/rcsb_pdb/mom_2011_12 10.2210/rcsb_pdb/mom_2011_12]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3M9S OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=3M9S FirstGlance]. <br> |
| - | </td></tr><tr><td class="sblockLbl"><b>[[ | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 4.5Å</td></tr> |
| - | + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=FES:FE2/S2+(INORGANIC)+CLUSTER'>FES</scene>, <scene name='pdbligand=FMN:FLAVIN+MONONUCLEOTIDE'>FMN</scene></td></tr> | |
| - | <tr><td class="sblockLbl"><b>[[ | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=3m9s FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3m9s OCA], [https://pdbe.org/3m9s PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=3m9s RCSB], [https://www.ebi.ac.uk/pdbsum/3m9s PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=3m9s ProSAT]</span></td></tr> |
| - | + | </table> | |
| - | <tr><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | + | == Function == |
| - | <table> | + | [https://www.uniprot.org/uniprot/NQO1_THET8 NQO1_THET8] NDH-1 shuttles electrons from NADH, via FMN and iron-sulfur (Fe-S) centers, to quinones in the respiratory chain. The immediate electron acceptor for the enzyme in this species is menaquinone. Couples the redox reaction to proton translocation (for every two electrons transferred, four hydrogen ions are translocated across the cytoplasmic membrane), and thus conserves the redox energy in a proton gradient required for the synthesis of ATP. The nqo1 subunit contains the NADH-binding site and the primary electron acceptor FMN. |
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
Check<jmol> | Check<jmol> | ||
<jmolCheckbox> | <jmolCheckbox> | ||
| - | <scriptWhenChecked>select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/m9/3m9s_consurf.spt"</scriptWhenChecked> | + | <scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/m9/3m9s_consurf.spt"</scriptWhenChecked> |
| - | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/ | + | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview03.spt</scriptWhenUnchecked> |
<text>to colour the structure by Evolutionary Conservation</text> | <text>to colour the structure by Evolutionary Conservation</text> | ||
</jmolCheckbox> | </jmolCheckbox> | ||
| - | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/ | + | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=3m9s ConSurf]. |
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
<div style="background-color:#fffaf0;"> | <div style="background-color:#fffaf0;"> | ||
| Line 25: | Line 26: | ||
The architecture of respiratory complex I.,Efremov RG, Baradaran R, Sazanov LA Nature. 2010 May 27;465(7297):441-5. PMID:20505720<ref>PMID:20505720</ref> | The architecture of respiratory complex I.,Efremov RG, Baradaran R, Sazanov LA Nature. 2010 May 27;465(7297):441-5. PMID:20505720<ref>PMID:20505720</ref> | ||
| - | From | + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> |
</div> | </div> | ||
| + | <div class="pdbe-citations 3m9s" style="background-color:#fffaf0;"></div> | ||
| + | |||
| + | ==See Also== | ||
| + | *[[NADH-quinone oxidoreductase|NADH-quinone oxidoreductase]] | ||
== References == | == References == | ||
<references/> | <references/> | ||
| Line 32: | Line 37: | ||
</StructureSection> | </StructureSection> | ||
[[Category: Complex I]] | [[Category: Complex I]] | ||
| + | [[Category: Large Structures]] | ||
[[Category: RCSB PDB Molecule of the Month]] | [[Category: RCSB PDB Molecule of the Month]] | ||
| - | [[Category: Thermus thermophilus]] | + | [[Category: Thermus thermophilus HB8]] |
| - | [[Category: Baradaran | + | [[Category: Baradaran R]] |
| - | [[Category: Efremov | + | [[Category: Efremov RG]] |
| - | [[Category: Sazanov | + | [[Category: Sazanov LA]] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
Crystal structure of respiratory complex I from Thermus thermophilus
| |||||||||||