We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox 126
From Proteopedia
(Difference between revisions)
| (2 intermediate revisions not shown.) | |||
| Line 12: | Line 12: | ||
==='''Catalytic Mechanism of Action of Transpeptidases'''=== | ==='''Catalytic Mechanism of Action of Transpeptidases'''=== | ||
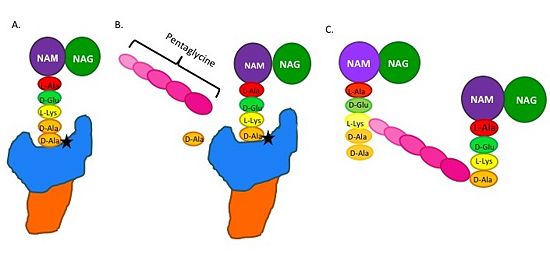
(A) The peptidoglycan D-Ala D-Ala moiety enters the TP active site, which is in the TP domain (blue). | (A) The peptidoglycan D-Ala D-Ala moiety enters the TP active site, which is in the TP domain (blue). | ||
| + | |||
(B) The active site serine residue (star) reacts with and breaks the peptide bond between the D-Ala residues. The terminal D-Ala residue exits the active site. The remaining D-Ala residue is covalently bound to the active site serine residue, and therefore, to TP. The incoming pentaglycine chain reacts with the bound D-Ala residue and is cross-linked to the D-Ala residue. | (B) The active site serine residue (star) reacts with and breaks the peptide bond between the D-Ala residues. The terminal D-Ala residue exits the active site. The remaining D-Ala residue is covalently bound to the active site serine residue, and therefore, to TP. The incoming pentaglycine chain reacts with the bound D-Ala residue and is cross-linked to the D-Ala residue. | ||
(C) This results in cross-linking between adjacent peptidoglycan "sheets" and regeneration of the active site serine residue so it can catalyze another cross-linking reaction.[[Image: MechanismofPBP.jpg|thumb|alt= Alt text|Figure 2.Schematic showing Catalytic Mechanism of PBP2a |550px]] | (C) This results in cross-linking between adjacent peptidoglycan "sheets" and regeneration of the active site serine residue so it can catalyze another cross-linking reaction.[[Image: MechanismofPBP.jpg|thumb|alt= Alt text|Figure 2.Schematic showing Catalytic Mechanism of PBP2a |550px]] | ||
| Line 23: | Line 24: | ||
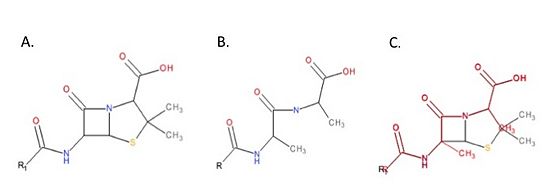
MRSA becomes resistant to beta-lactams by acquiring an alternative TP, PBP2a, that is encoded by the ''mecA'' gene (Matsuhashi ''et al.'', 1986). PBP2a is compromised in its ability to react with beta-lactam; therefore, MRSA strains are resistant to beta-lactams and are able to make their cell wall in the presence of high concentrations of beta-lactams. | MRSA becomes resistant to beta-lactams by acquiring an alternative TP, PBP2a, that is encoded by the ''mecA'' gene (Matsuhashi ''et al.'', 1986). PBP2a is compromised in its ability to react with beta-lactam; therefore, MRSA strains are resistant to beta-lactams and are able to make their cell wall in the presence of high concentrations of beta-lactams. | ||
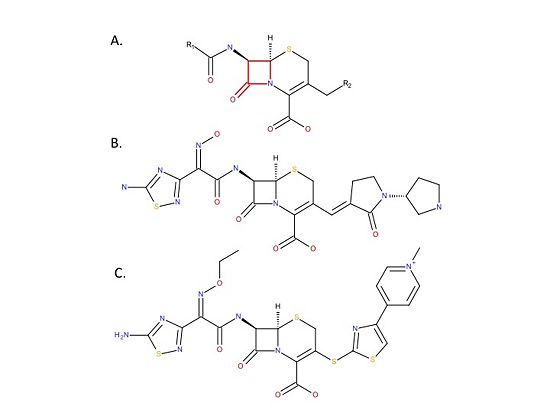
| - | Recently, two broad range cephalosporins: ceftaroline and ceftaroline (Figure), that have anti-MRSA activity because they bind and inhibit PBP2a have been developed. | + | Recently, two broad range cephalosporins: ceftaroline and ceftaroline (Figure 4), that have anti-MRSA activity because they bind and inhibit PBP2a have been developed.[[Image:MedicinesSchematic.jpg|thumb|alt= Alt text| Figure 4. Chemical structure of anti-MRSA cephalosporins. A. Chemical structure of the cephalosporin backbone. Cephalosporins are beta-lactam antibiotics that have a core backbone that includes a beta-lactam ring (red). There are many different cephalosporins, all of which have different antimicrobial activities and chemical properties. The differences between the cephalosporins is due to differences in the R1 and R2 groups. B. Chemical structure of ceftobiprole. C. Chemical structure of ceftaroline. |550px]] |
Current revision
| |||||||||||