Journal:JBIC:26
From Proteopedia
(Difference between revisions)

| (4 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
<StructureSection load='' size='450' side='right' scene='59/596313/Cv/15' caption=''> | <StructureSection load='' size='450' side='right' scene='59/596313/Cv/15' caption=''> | ||
=== Fluoride inhibition of Sporosarcina pasteurii urease: structure and thermodynamics === | === Fluoride inhibition of Sporosarcina pasteurii urease: structure and thermodynamics === | ||
| - | <big>Stefano Benini, Michele Cianci, Luca Mazzei and Stefano Ciurli</big> <ref> | + | <big>Stefano Benini, Michele Cianci, Luca Mazzei and Stefano Ciurli</big> <ref>PMID 25113581 </ref> |
<hr/> | <hr/> | ||
<b>Molecular Tour</b><br> | <b>Molecular Tour</b><br> | ||
| Line 7: | Line 7: | ||
[[Image:Scheme_1.png|left|450px|thumb|]] | [[Image:Scheme_1.png|left|450px|thumb|]] | ||
{{Clear}} | {{Clear}} | ||
| - | A strategy to control the activity of urease for medical and agricultural applications is to use enzyme inhibitors. Fluoride is a known urease inhibitor, but the structural basis of its mode of inhibition are still undetermined. Here, kinetic studies on the fluoride-induced inhibition of urease from ''Sporosarcina pasteurii'', a widespread and highly ureolytic soil bacterium, revealed a mixed competitive and uncompetitive mechanism. The pH-dependence of the inhibition constants, investigated in the 6.5-8.0 range, reveals a predominant uncompetitive mechanism that increases by increasing the pH, and a lesser competitive inhibition that increases by lowering the pH. Ten crystal structures of the enzyme were independently determined using five crystals of the <scene name='59/596313/Cv/13'>native form</scene> and five crystals of the protein crystallised in the presence of fluoride. The analysis of these structures revealed the presence of <scene name='59/596313/Cv/14'>two fluoride anions coordinated to the Ni(II) ions in the active site</scene>, in terminal and bridging positions (<span style="color:gold;background-color:black;font-weight:bold;"> | + | A strategy to control the activity of urease for medical and agricultural applications is to use enzyme inhibitors. Fluoride is a known urease inhibitor, but the structural basis of its mode of inhibition are still undetermined. Here, kinetic studies on the fluoride-induced inhibition of urease from ''Sporosarcina pasteurii'', a widespread and highly ureolytic soil bacterium, revealed a mixed competitive and uncompetitive mechanism. The pH-dependence of the inhibition constants, investigated in the 6.5-8.0 range, reveals a predominant uncompetitive mechanism that increases by increasing the pH, and a lesser competitive inhibition that increases by lowering the pH. Ten crystal structures of the enzyme were independently determined using five crystals of the <scene name='59/596313/Cv/13'>native form</scene> and five crystals of the protein crystallised in the presence of fluoride. The analysis of these structures revealed the presence of <scene name='59/596313/Cv/14'>two fluoride anions coordinated to the Ni(II) ions in the active site</scene>, in terminal and bridging positions (<span style="color:gold;background-color:black;font-weight:bold;">both fluorides are colored in gold</span>). <scene name='59/596313/Cv/20'>Click here to see animation</scene>. |
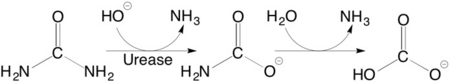
Structural studies on ureases have revealed that the immediate environment around the two Ni(II) ions at the active site is conserved, as to induce a common mechanism of catalysis whose key step is the nucleophilic attack of the nickel-bridging hydroxide on the urea molecule bound to the bimetallic nickel cluster via O and N atoms (see static image below). | Structural studies on ureases have revealed that the immediate environment around the two Ni(II) ions at the active site is conserved, as to induce a common mechanism of catalysis whose key step is the nucleophilic attack of the nickel-bridging hydroxide on the urea molecule bound to the bimetallic nickel cluster via O and N atoms (see static image below). | ||
[[Image:Scheme_2.png|left|450px|thumb|]] | [[Image:Scheme_2.png|left|450px|thumb|]] | ||
{{Clear}} | {{Clear}} | ||
| - | The present study consistently supports an interaction of fluoride with the nickel centres in the urease active site in which <scene name='59/596313/Cv/17'>one fluoride competitively binds</scene> to the Ni(II) ion proposed to coordinate urea in the initial step of the catalytic mechanism, while <scene name='59/596313/Cv/18'>another fluoride uncompetitively substitutes</scene> the Ni(II)-bridging hydroxide, blocking its nucleophilic attack on urea. | + | The present study consistently supports an interaction of fluoride with the nickel centres in the urease active site in which <scene name='59/596313/Cv/17'>one fluoride competitively binds</scene> (<span style="color:salmon;background-color:black;font-weight:bold;">colored in salmon</span>) to the Ni(II) ion proposed to coordinate urea in the initial step of the catalytic mechanism, while <scene name='59/596313/Cv/18'>another fluoride uncompetitively substitutes</scene> (<span style="color:cyan;background-color:black;font-weight:bold;">colored in cyan</span>) the Ni(II)-bridging hydroxide, blocking its nucleophilic attack on urea. |
| + | |||
| + | '''PDB references:''' 1.58 A resolution native ''Sporosarcina pasteurii'' urease [[4ceu]]; 1.59 A resolution Fluoride inhibited ''Sporosarcina pasteurii'' urease [[4cex]]. | ||
</StructureSection> | </StructureSection> | ||
<references/> | <references/> | ||
__NOEDITSECTION__ | __NOEDITSECTION__ | ||
Current revision
| |||||||||||
- ↑ Benini S, Cianci M, Mazzei L, Ciurli S. Fluoride inhibition of Sporosarcina pasteurii urease: structure and thermodynamics. J Biol Inorg Chem. 2014 Aug 12. PMID:25113581 doi:http://dx.doi.org/10.1007/s00775-014-1182-x
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.