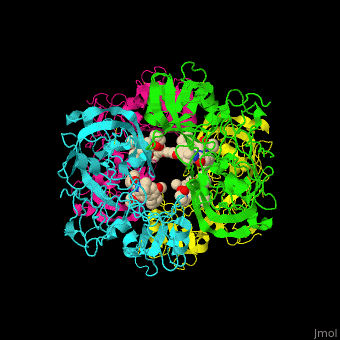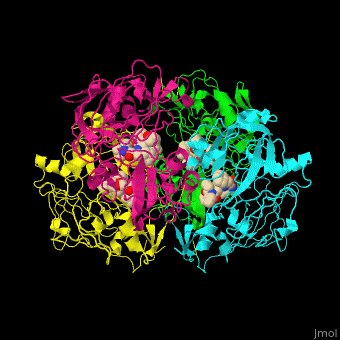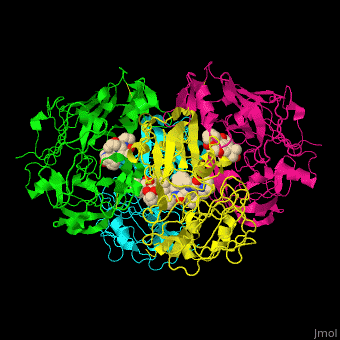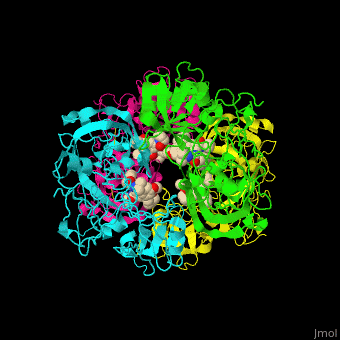
Plasmepsin
From Proteopedia
(Difference between revisions)
| (18 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='' size='350' side='right' caption='Plasmepsin II complex with a potential antimalarial drug (PDB entry [[4cku]])' scene='44/441867/Cv/1'> | |
| - | [[Plasmepsin]] (Plm) is a hemoglobin-degrading enzyme produced by the plasmodium parasite | + | == Function == |
| + | [[Plasmepsin]] (Plm) is a hemoglobin-degrading enzyme produced by the plasmodium parasite. It is an aspartic acid protease having 2 aspartic acid residues in the active site. Ten Plm isoforms are known which are named Plm I, II, etc and '''Histo-Aspartic Protease (HAP)'''. | ||
| - | < | + | *'''Proplasmepsin II''' exhibits a large shift between its domains which renders the protease inactive<ref>PMID:9886289</ref>. |
| + | ==Relevance == | ||
| + | Plm is a potential target for anti-malaria drugs<ref>PMID:25719272</ref>. | ||
| + | |||
| + | == Structural highlights == | ||
| + | The active site of Plm II contains a <scene name='44/441867/Cv/4'>catalytic dyad of 2 Asp residues which interact with potential antimalarial inhibitor</scene><ref>PMID:24900843</ref>. <scene name='44/441867/Cv/5'>Whole active site</scene>. Water molecules are shown as red spheres. | ||
| + | </StructureSection> | ||
==3D structures of plasmepsin== | ==3D structures of plasmepsin== | ||
| - | Updated | + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} |
| + | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| + | |||
| + | *Histo-Aspartic Protease | ||
| + | |||
| + | **[[3fns]], [[3qvc]], [[6kub]], [[6kuc]], [[6kud]] – PfHAP - ''Plasmodium falciparum''<br /> | ||
| + | **[[3fnt]] – PfHAP + pepstatin<br /> | ||
| + | **[[3fnu]], [[3qvi]] – PfHAP + inhibitor<br /> | ||
| + | |||
| + | *Plasmepsin I | ||
| - | + | **[[3qrv]] – PfPlm I residues 117-457<br /> | |
| + | **[[3qs1]] - PfPlm I residues 117-457 + inhibitor | ||
| - | + | *Plasmepsin II | |
| - | + | ||
| - | + | ||
| - | + | **[[1lf4]] – PfPlm II<br /> | |
| + | **[[3f9q]] – PfPlm II (mutant)<br /> | ||
| + | **[[2r9b]] – PfPlm II + peptide inhibitor<br /> | ||
| + | **[[1w6h]], [[2bju]], [[1lf2]], [[1lf3]], [[1lee]], [[2igx]], [[2igy]], [[4cku]], [[4y6m]], [[4z22]], [[5yia]], [[5yib]], [[5yic]], [[5yid]], [[5yie]], [[7qyh]] – PfPlm II + inhibitor<br /> | ||
| + | **[[4ya8]] – PfPlm II (mutant) + inhibitor<br /> | ||
| + | **[[7ve0]], [[7ve2]] – PfPlm II + drug<br /> | ||
| + | **[[1w6i]], [[1xdh]], [[1xe5]], [[1xe6]], [[1me6]], [[1sme]] – PfPlm II + pepstatin derivative<br /> | ||
| + | **[[1m43]] - PfPlm II (mutant) + pepstatin derivative<br /> | ||
| - | + | *Plasmepsin IV | |
| - | + | ||
| - | + | **[[2anl]] – Plm IV + statine derivative – ''Plasmodium malariae''<br /> | |
| + | **[[1ls5]] - PfPlm IV + pepstatin derivative<br /> | ||
| - | + | *Plasmepsin V or aspartic protease PM5 | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | **[[6c4g]] – PvPlm V + inhibitor<br /> | |
| + | **[[4zl4]] – PvPlm V + transition state analog<br /> | ||
| - | + | *Plasmepsin X | |
| - | + | ||
| - | + | ||
| - | + | **[[7ry7]] – PvPlm X <br /> | |
| + | **[[7tbc]], [[7tbd]], [[7tbe]], [[8dsr]] – PfPlm X + inhibitor<br /> | ||
| + | **[[7tbb]] – PfPlm X <br /> | ||
| - | + | *Proplasmepsin II | |
| - | + | ||
| + | **[[1pfz]], [[5bwy]] - PfProPlm II<br /> | ||
| + | **[[1miq]] – PvProPlm II – ''Plasmodium vivax''<br /> | ||
| + | **[[1qs8]], [[5i70]] - PvPlm II + pepstatin derivative<br /> | ||
| + | }} | ||
| + | == References == | ||
| + | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
3D structures of plasmepsin
Updated on 31-August-2023
References
- ↑ Bernstein NK, Cherney MM, Loetscher H, Ridley RG, James MN. Crystal structure of the novel aspartic proteinase zymogen proplasmepsin II from plasmodium falciparum. Nat Struct Biol. 1999 Jan;6(1):32-7. PMID:9886289 doi:10.1038/4905
- ↑ Huizing AP, Mondal M, Hirsch AK. Fighting malaria: structure-guided discovery of nonpeptidomimetic plasmepsin inhibitors. J Med Chem. 2015 Jul 9;58(13):5151-63. doi: 10.1021/jm5014133. Epub 2015 Mar 17. PMID:25719272 doi:http://dx.doi.org/10.1021/jm5014133
- ↑ Jaudzems K, Tars K, Maurops G, Ivdra N, Otikovs M, Leitans J, Kanepe-Lapsa I, Domraceva I, Mutule I, Trapencieris P, Blackman MJ, Jirgensons A. Plasmepsin inhibitory activity and structure-guided optimization of a potent hydroxyethylamine-based antimalarial hit. ACS Med Chem Lett. 2014 Jan 13;5(4):373-7. doi: 10.1021/ml4004952. eCollection, 2014 Apr 10. PMID:24900843 doi:http://dx.doi.org/10.1021/ml4004952
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Joel L. Sussman, Jaime Prilusky