Sigma factor
From Proteopedia
(Difference between revisions)
| (13 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
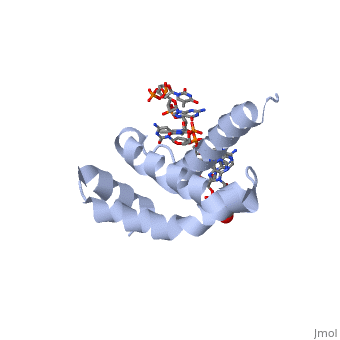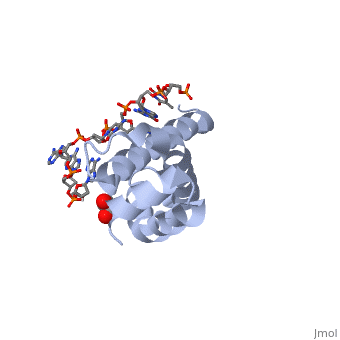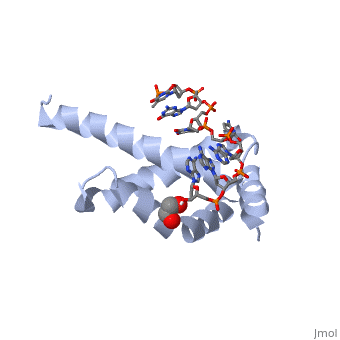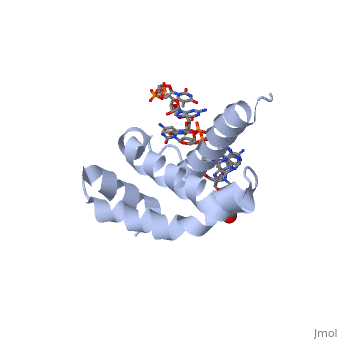
| + | <StructureSection load='4lup' size='340' side='right' caption='Structure of sigma factor of E.Coli RNAP (grey) in complex with promoter DNA and ethylene glycol (PDB code [[4lup]]).' scene=''> | ||
| - | <StructureSection load='4lup' size='340' side='right' caption='Structure of sigma factor of E.Coli RNAP in complex with Promoter DNA (PDB code [[4lup]]).' scene=''> | ||
| - | |||
| - | [[Image:2h27.jpg|left|200px]] | ||
== Overview == | == Overview == | ||
| - | '''Sigma (σ) factor''' is the peoptide subunit needed for the initiation of RNA transcription in prokaryotic organisms <scene name='59/591940/Sigma_factor_in_enzyme/1'>as seen here</scene>. As opposed to eukaryotes, who utilize a variety of proteins to initiate gene transcription, prokaryotic transcription is initiated almost completely by a σ-factor. The large and biologically essential protein, RNA polymerase (RNAP), contains one σ-subunit, which binds <scene name='59/591940/Dna_promoter/3'>DNA promoter sequences</scene>, located upstream of transcription start sites. | + | '''Sigma (σ) factor''' is the peoptide subunit needed for the initiation of RNA transcription in prokaryotic organisms <scene name='59/591940/Sigma_factor_in_enzyme/1'>as seen here</scene>. As opposed to eukaryotes, who utilize a variety of proteins to initiate gene transcription, prokaryotic transcription is initiated almost completely by a σ-factor. The large and biologically essential protein, RNA polymerase (RNAP), contains one σ-subunit, which binds <scene name='59/591940/Dna_promoter/3'>DNA promoter sequences</scene>, located upstream of transcription start sites. <br /> |
| + | *'''Sigma factor 70''' or '''RpoD''' is the primary initiation factor during exponential growth. <br /> | ||
| + | *'''Sigma factor 45''' or '''RpoN''' is responsible for the expression of genes involved in Arg catabolism.<br /> | ||
| + | *'''Sigma factor 38''' or '''RpoS''' acts as the master regulator of the general stress response in ''E. coli'' .<br /> | ||
== Function and Structure == | == Function and Structure == | ||
| - | The '''σ-factor''' performs two chief functions: to direct the catalytic core of RNAP to the promotoer upstream of the +1 start site of transcription, and finally to assist in the initiation of strand seperation of double-helical DNA, forming the transcription "bubble." Each gene promoter utilizes a specific promoter region about 40 bp upstream of the transcription start site, and therefore different σ-factors play a role in the regulation of different genes. This process, which includes association of the σ-factor with RNAP to recognize and open DNA at the promoter site, followed by dissociation of the σ to allow elongation, which can then activate additional RNAP enzymes, is referred to as the '''σ-cycle'''. | + | The '''σ-factor''' performs two chief functions: to direct the catalytic core of RNAP to the promotoer upstream of the +1 start site of transcription, and finally to assist in the initiation of strand seperation of double-helical DNA, forming the transcription "bubble."[1] Each gene promoter utilizes a specific promoter region about 40 bp upstream of the transcription start site, and therefore different σ-factors play a role in the regulation of different genes [2]. This process, which includes association of the σ-factor with RNAP to recognize and open DNA at the promoter site, followed by dissociation of the σ to allow elongation, which can then activate additional RNAP enzymes, is referred to as the '''σ-cycle''' [3]. |
===Domain Strucure & DNA interactions=== | ===Domain Strucure & DNA interactions=== | ||
| - | There are many types of σ-subunits, and each recognizes a unique promoter sequence. Furthmore, each unique σ is composed of a variable number of structured domains. The simplest σ-factors have two domains, few have three, and most, called '''housekeeping σ-factors''', have 4 domains, given the names σ(4), σ(3), σ(2), and σ(1.1). All domains are linked by very flexible peptide '''linkers''' which can extend very long distances. Each of these domains utilizes DNA-binding determinants, or domains that recognize specific sequences and conformations in DNA. Most commonly, these recognized sequences occur at the -35 and -10 locations upstream of the +1 site. One such DNA-binding motif, '''the helix-turn-helix motif''' (<scene name='59/591940/Hth_motif/2'>HTH</scene>), helps specifically recognize DNA promoters at both the -35 and -10 positions. This HTH motif, used by most σ-factors, maintains its specificity and accuracy by binding in the '''major groove''' of DNA, where it can interact with the base pairs in the DNA double-helix. In many prokaryotes, these portions of DNA maintain consensus adenosine and thymine sequences, such as <scene name='59/591940/Ta_sequence/1'>TATAAT</scene>. | + | There are many types of σ-subunits, and each recognizes a unique promoter sequence. Furthmore, each unique σ is composed of a variable number of structured domains. The simplest σ-factors have two domains, few have three, and most, called '''housekeeping σ-factors''', have 4 domains, given the names σ(4), σ(3), σ(2), and σ(1.1) [1,3]. All domains are linked by very flexible peptide '''linkers''' which can extend very long distances. Each of these domains utilizes DNA-binding determinants, or domains that recognize specific sequences and conformations in DNA. Most commonly, these recognized sequences occur at the -35 and -10 locations upstream of the +1 site. One such DNA-binding motif, '''the helix-turn-helix motif''' (<scene name='59/591940/Hth_motif/2'>HTH</scene>), helps specifically recognize DNA promoters at both the -35 and -10 positions [1]. This HTH motif, used by most σ-factors, maintains its specificity and accuracy by binding in the '''major groove''' of DNA, where it can interact with the base pairs in the DNA double-helix. In many prokaryotes, these portions of DNA maintain consensus adenosine and thymine sequences [1,2], such as <scene name='59/591940/Ta_sequence/1'>TATAAT</scene>. |
===Transcription Bubble=== | ===Transcription Bubble=== | ||
| - | The <scene name='59/591940/Transcription_bubble/1'>transcription bubble</scene>, also referred to as the '''open complex''' is formed through the common '''housekeeping σ factors''' which unwind about 13 bp of duplex DNA in an ATP independent process. Research has shown that σ factors require invariant basic and aromatic residues (Phe, Tyr, Trp) critical for this formation. The process of bubble formation begins at the -11 formation (usually A) and propogates to +1 site, through a phenomenon called <scene name='59/591940/Transcription_bubble_flipped/1'>Base Flipping</scene>, which interrupts the stacking interactions stabilizing the double helix conformation. As this process occurs and the DNA transitions into the open promoter complex, certain RNAP-σ contacts are lost, initiating the dissociation of σ. In summary, the processes of -35 and -10 motif sequence recognition and helix strand separation are coupled by the σ factor. | + | The <scene name='59/591940/Transcription_bubble/1'>transcription bubble</scene>, also referred to as the '''open complex''' is formed through the common '''housekeeping σ factors''' which unwind about 13 bp of duplex DNA in an ATP independent process. Research has shown that σ factors require invariant basic and aromatic residues (Phe, Tyr, Trp) critical for this formation [1]. The process of bubble formation begins at the -11 formation (usually A) and propogates to +1 site, through a phenomenon called <scene name='59/591940/Transcription_bubble_flipped/1'>Base Flipping</scene>, which interrupts the stacking interactions stabilizing the double helix conformation [1]. As this process occurs and the DNA transitions into the open promoter complex, certain RNAP-σ contacts are lost, initiating the dissociation of σ. In summary, the processes of -35 and -10 motif sequence recognition and helix strand separation are coupled by the σ factor. |
==Restriction== | ==Restriction== | ||
| - | Initiation of prokaryotic transcription requires cooperation between the σ peptide and RNAP. Without these interactions, no transcription is possible. | + | Initiation of prokaryotic transcription requires cooperation between the σ peptide and RNAP. Without these fundamental interactions, no transcription is possible. |
===Comformational and Autoinhibitory=== | ===Comformational and Autoinhibitory=== | ||
| - | Normally, σ-factor domains cannot bind to promoters on their own. These domains usually are placed in very compacted positions relative to each other, a conformation that buries DNA-binding determinants. This type of restriction is called '''conformational restriction'''. Additionally, in housekeeping σs, a domain called the '''σ(1.1)''' stabilizes the compact conformation mentioned above, thereby preventing any promoter recognition. This method of restricting the binding abilities of isolated σ's is called '''autoinhibitory inhibition'''. | + | Normally, σ-factor domains cannot bind to promoters on their own. These domains usually are placed in very compacted positions relative to each other, a conformation that buries DNA-binding determinants. This type of restriction is called '''conformational restriction'''[1]. Additionally, in housekeeping σs, a domain called the '''σ(1.1)''' stabilizes the compact conformation mentioned above, thereby preventing any promoter recognition. This method of restricting the binding abilities of isolated σ's is called '''autoinhibitory inhibition'''[1]. |
===anti-σ's=== | ===anti-σ's=== | ||
An additional method of restriction is through the action of '''anti-σ's''', which act by making stable interactions with σ-domains, such as σ | An additional method of restriction is through the action of '''anti-σ's''', which act by making stable interactions with σ-domains, such as σ | ||
| - | (4), which allows them to make energy-favorable interactions with RNAP residues. This causes a cascading "peeling off" effect of other σ-domains from the RNAP, preventing any interaction with duplex DNA and inhibiting transcription in an analogous process to competitive inhibition. | + | (4), which allows them to make energy-favorable interactions with RNAP residues. This causes a cascading "peeling off" effect of other σ-domains from the RNAP, preventing any interaction with duplex DNA and inhibiting transcription in an analogous process to competitive inhibition [3]. |
== Gene Regulation and Differentiation == | == Gene Regulation and Differentiation == | ||
| - | Since σ-factors are exclusively linked to gene expression in prokaryotic organisms, the variety of σ-factors in a cell dictate how and what genes are transcribed. Specialized function in cells, therefore, is highly moderated by its arsenal of σ-subunits. In fact, cellular development and differentiation are directly impacted and carried out by "cascades" of σ-factors. In the early stages of development, '''early genes''' are transcribed by basic '''bacterial σ-factors'''. These genes are therefore transcribed to give new σ-factors, which in turn activate additional genes, and so on. This process of σ-factor cascades demonstrates the versatile and essential biologic functions of the RNAP subunit, σ. | + | Since σ-factors are exclusively linked to gene expression in prokaryotic organisms, the variety of σ-factors in a cell dictate how and what genes are transcribed. Specialized function in cells, therefore, is highly moderated by its arsenal of σ-subunits. In fact, cellular development and differentiation are directly impacted and carried out by "cascades" of σ-factors. In the early stages of development, '''early genes'''[2] are transcribed by basic '''bacterial σ-factors'''. These genes are therefore transcribed to give new σ-factors, which in turn activate additional genes, and so on [2]. This process of σ-factor cascades demonstrates the versatile and essential biologic functions of the RNAP subunit, σ. |
| - | + | ||
==3D structures of sigma factor== | ==3D structures of sigma factor== | ||
| + | [[Sigma factor 3D structures]] | ||
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
== References == | == References == | ||
| - | Felklistov, Andrey, Brian D. Sharon, Seth A. Darst, and Carol A. Gross. "Bacterial Sigma Factors: A Historical, Structural, and Genomic Perspective." The Annual Review of Microbiology 68 (2014): 357-76. | + | 1. Felklistov, Andrey, Brian D. Sharon, Seth A. Darst, and Carol A. Gross. "Bacterial Sigma Factors: A Historical, Structural, and Genomic Perspective." The Annual Review of Microbiology 68 (2014): 357-76. |
| - | Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. 3rd ed. Hoboken, NJ: Wiley, 2008. | + | 2. Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. 3rd ed. Hoboken, NJ: Wiley, 2008. |
| - | Mooney, R. A., S. A. Darst, and R. Landick. "Sigma and RNA Polymerase: An On-again, Off-again Relationship?" Molecular Cell 20.3 (2005): 335-45. | + | 3. Mooney, R. A., S. A. Darst, and R. Landick. "Sigma and RNA Polymerase: An On-again, Off-again Relationship?" Molecular Cell 20.3 (2005): 335-45. |
<references/> | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
References
1. Felklistov, Andrey, Brian D. Sharon, Seth A. Darst, and Carol A. Gross. "Bacterial Sigma Factors: A Historical, Structural, and Genomic Perspective." The Annual Review of Microbiology 68 (2014): 357-76.
2. Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. 3rd ed. Hoboken, NJ: Wiley, 2008.
3. Mooney, R. A., S. A. Darst, and R. Landick. "Sigma and RNA Polymerase: An On-again, Off-again Relationship?" Molecular Cell 20.3 (2005): 335-45.