Molecular Playground/E. coli ClpP
From Proteopedia
(Difference between revisions)
| (110 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | Here, in the Chien lab of the University of Massachusetts-Amherst, we study how [http://en.wikipedia.org/wiki/Proteolysis proteolysis] plays a large part in protein quality control. The maintenance and timely destruction of protein levels plays an important role during cell homeostasis and cell transitions/differentiation, yet much of what governs these processes has to be fully understood. | |
| - | + | ||
| - | + | ||
<StructureSection load='1yg6' size='340' side='right' caption='Caption for this structure' scene=''> | <StructureSection load='1yg6' size='340' side='right' caption='Caption for this structure' scene=''> | ||
| - | This is a default text for your page '''Lisa Hernandez/Sandbox1'''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
| - | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
| - | |||
| - | == Function == | ||
| - | |||
| - | == Disease == | ||
| - | |||
| - | == Relevance == | ||
| + | == ClpP Introduction == | ||
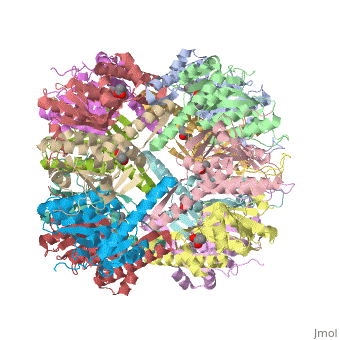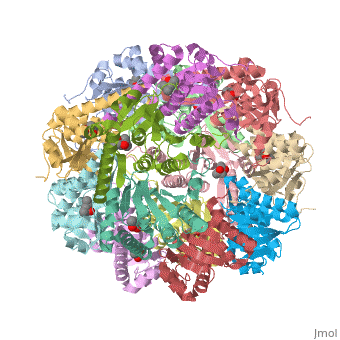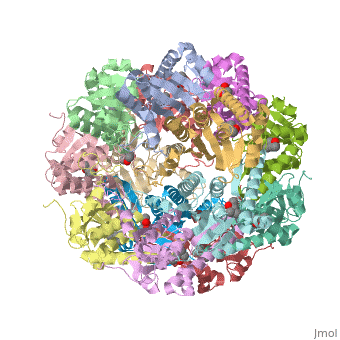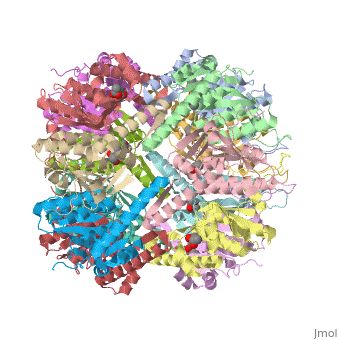
| + | ''E. coli'' Casein lytic proteinase P (<scene name='60/609790/Clpp-2/2'>ClpP</scene>) is a double ring [http://en.wiktionary.org/wiki/tetradecamer tetradecameric] homo oligomer compartmentalized peptidase [http://www.ncbi.nlm.nih.gov/pubmed/?term=ClpP%3A+A+structurally+dynamic+protease+regulated+by+AAA%2B+proteins]. ClpP requires the use of ATP dependent regulatory elements that independently bind to ClpP in order for substrates to have access the active core [http://www.ncbi.nlm.nih.gov/pubmed/15037252][http://www.ncbi.nlm.nih.gov/pubmed/20236930][http://www.ncbi.nlm.nih.gov/pubmed/19038348]. Here, proteins that are translocated by regulatory elements into the peptidase core are cleaved into smaller amino acid chains approximately on average 6-8aa in length [http://www.ncbi.nlm.nih.gov/pubmed/16229481]. | ||
| + | == Tetradecameric Structure == | ||
| + | ClpP is a serine protease which consists of fourteen monomers situated into two [http://en.wiktionary.org/wiki/heptamer heptameric] rings seated on top of each other. In the center of the barrel-shaped chamber lies a core of fourteen peptide-cleaving active sites, restricted by the narrow entrance called the <scene name='60/609790/Clpp_axial_pore/4'>axial pore</scene>. As peptides are [http://en.wikipedia.org/wiki/Processivity processively] threaded in the pore by the regulatory elements, the peptides are then degraded into smaller fragments as a result from ClpP cleavage. Smaller peptides are then released through small openings found around the <scene name='60/609790/Clpp_equator_pore/2'>equatorial interface</scene> of the two stacked rings. | ||
== Structural highlights == | == Structural highlights == | ||
| + | Featured within the core of ClpP are the catalytic sets of amino acids [X-Y-Z] that constitute the active mechanism featured by serene proteases [http://www.ncbi.nlm.nih.gov/pubmed/2197276][http://www.ncbi.nlm.nih.gov/pubmed/17499722]. Human ClpP can exist in two forms, a single heptameric ring or as a double stack set of hepatmeric rings where the double stacked form (tetradecamer) is the active form [http://www.ncbi.nlm.nih.gov/pubmed/16115876]. Tetradecamer ClpP is a stable but not a rigid structure that can undergo several conformational forms when interacting with its regulatory elements. By doing so both stabilizes processivity of translocation by the regulatory elements and is thought to increase likelihood of exposure to the active sites resulting in timely, small peptide formation. | ||
| - | + | == Role of ClpP/Biological Relevance == | |
| + | ''E. coli'' ClpAP/ClpXP complexes play a critical role in maintaining protein homeostasis under several levels of quality control. Improperly folded or aggregated proteins are potential ClpP substrates based on properties of the associated regulatory element recognition. Targeted removal of aberrant proteins resulting from and rescue of stalled ribosomes by the SsrA tagging system are directly recognized and degraded by ClpAP/ClpXP complexes [http://www.ncbi.nlm.nih.gov/pubmed/9573050]. In ''E. coli'' ClpP and ClpP homologues found in other bacteria require regulatory elements to recognize and import proteins for destruction. To gain access to the active sites is tightly controlled and therefore a potential antimicrobial target where loss of regulation (for example, through use of acyldepsipeptides or ADEPs) literally digests the bacteria from the inside out [http://link.springer.com/chapter/10.1007/978-94-007-6787-4_24]. ''E. coli'' ClpAP and ClpXP has been used as a structural model for the 26 proteasome to gain insight into its workings [http://www.ncbi.nlm.nih.gov/pubmed/7623377][http://www.ncbi.nlm.nih.gov/pubmed/14990998]. | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
| + | [http://www.ncbi.nlm.nih.gov/pubmed/?term=ClpP%3A+A+structurally+dynamic+protease+regulated+by+AAA%2B+proteins 1. ''ClpP: A structurally dynamic protease regulated by AAA+ proteins.'' Alexopoulos JA ''et. al'' (2012 J Struct Bio)] | ||
| + | |||
| + | [http://www.ncbi.nlm.nih.gov/pubmed/15037252 2. ''ClpA and ClpX ATPases bind simultaneously to opposite ends of ClpP peptidase to form active hybrid complexes.'' Ortega J ''et. al'' (2004 J. Struct Biol.)] | ||
| + | |||
| + | [http://www.ncbi.nlm.nih.gov/pubmed/20236930 3. ''Binding of the ClpA Unfoldase Opens the Axial Gate of ClpP Peptidase.'' Effantin ''et. al'' (2010 J Biol Chem)] | ||
| + | |||
| + | [http://www.ncbi.nlm.nih.gov/pubmed/19038348 4. ''Turned on for degradation: ATPase-independent degradation by ClpP.'' Bewley MC ''et. al'' (2009 J Struct Biol)] | ||
| + | |||
| + | [http://www.ncbi.nlm.nih.gov/pubmed/16229481 5. ''Control of peptide product sizes by the energy-dependent protease ClpAP.'' Choi KH ''et. al'' (2005 Biochemistry)] | ||
| + | |||
| + | [http://www.ncbi.nlm.nih.gov/pubmed/2197276 6. ''Clp P represents a unique family of serine proteases.'' Maurizi MR ''et. al'' (1990 J Biolchem)] | ||
| + | |||
| + | [http://www.ncbi.nlm.nih.gov/pubmed/17499722 7. ''ClpP: a distinctive family of cylindrical energy-dependent serine proteases.'' Yu AY ''et. al'' (2007 FEBS Lett.)] | ||
| + | |||
| + | [http://www.ncbi.nlm.nih.gov/pubmed/16115876 8. ''Human mitochondrial ClpP is a stable heptamer that assembles into a tetradecamer in the presence of ClpX.'' Kang SG ''et. al'' (2007 J Biol Chem)] | ||
| + | |||
| + | [http://www.ncbi.nlm.nih.gov/pubmed/9573050 9. ''The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system.'' Gottesman S ''et. al'' (1998 Genes Dev.)] | ||
| + | |||
| + | [http://link.springer.com/chapter/10.1007/978-94-007-6787-4_24 10. ''Bacterial Cell Stress Protein ClpP: A Novel Antibiotic Target.'' Brötz-Oesterhelt ''et. al'' (2013 Heat Shock Proteins Volume 7, pp 375-385)] | ||
| + | |||
| + | [http://www.ncbi.nlm.nih.gov/pubmed/7623377 11. ''Homology in structural organization between E. coli ClpAP protease and the eukaryotic 26 S proteasome.'' Kessel M ''et. al'' (1995 J Mol Bio)] | ||
| + | |||
| + | [http://www.ncbi.nlm.nih.gov/pubmed/14990998 12. ''Proteasomes and their kin: proteases in the machine age.'' Pickart CM ''et. al'' (2004 Nat Rev Mol Cell Biol.)] | ||
| + | |||
| + | |||
| + | == Acknowledgements== | ||
| + | Kamal Joshi, Joanne Lau, Jing Liu, Rob Vass | ||
| + | |||
| + | PDB id:1YG6 from [http://www.rcsb.org/pdb/explore.do?structureId=1yg6 Bewley, MC ''et. al'' (2006 J.Struct.Biol.)] | ||
Current revision
Here, in the Chien lab of the University of Massachusetts-Amherst, we study how proteolysis plays a large part in protein quality control. The maintenance and timely destruction of protein levels plays an important role during cell homeostasis and cell transitions/differentiation, yet much of what governs these processes has to be fully understood.
| |||||||||||
References
6. Clp P represents a unique family of serine proteases. Maurizi MR et. al (1990 J Biolchem)
Acknowledgements
Kamal Joshi, Joanne Lau, Jing Liu, Rob Vass
PDB id:1YG6 from Bewley, MC et. al (2006 J.Struct.Biol.)