We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Exosome
From Proteopedia
(Difference between revisions)
| (12 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
==Exosome== | ==Exosome== | ||
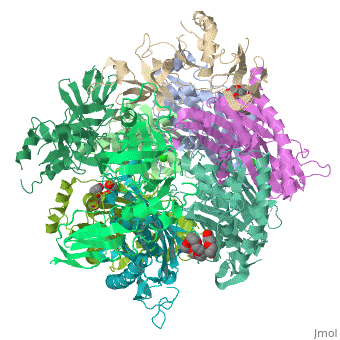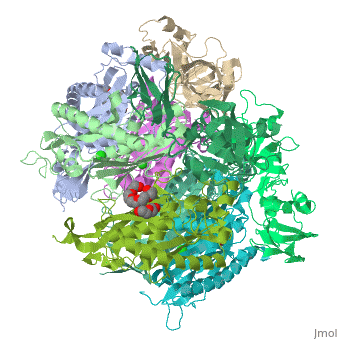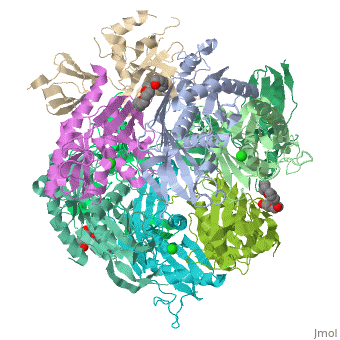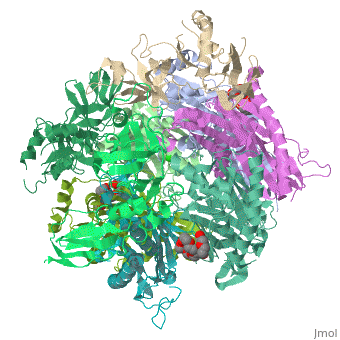
| - | <StructureSection load=' | + | <StructureSection load='2je6' size='340' side='right' caption='Exosome complex containing exonuclease 1 (green), exonuclease 2 (grey) and exosome complex RNA-binding protein 1 (wheat) complex with PEG400 (pdb code [[2je6]]).' scene=''> |
| - | The Exosome complex (or just- Exosome) is a multi-protein complex capable of degrading various types of RNA molecules. The Exosome complex is found in eukaryotic cell, and also in archaea, while in bacteria it is found as a simpler complex (but it has the same function). <ref>http://en.wikipedia.org/wiki/Exosome_complex#Structure</ref> | + | The '''Exosome complex''' (or just- Exosome) is a multi-protein complex capable of degrading various types of RNA molecules. The Exosome complex is found in eukaryotic cell, and also in archaea, while in bacteria it is found as a simpler complex (but it has the same function). <ref>http://en.wikipedia.org/wiki/Exosome_complex#Structure</ref> |
| + | |||
| + | Some of the exosome components are:<br /> | ||
| + | *'''MTR3''' and '''SKI6''' are part of RNase PH domain-containing subunits proposed to form a central channel which threads RNA for degradation.<br /> | ||
| + | *'''MTR4''' interacts with Nop53 and together they have a role in the maturation of 5.8S rRNA<ref>PMID:28883156</ref>.<br /> | ||
| + | *'''Rrp6''' is a nuclear-specific subunit promoting cell survival during heat stress<ref>PMID:32521279</ref>.<br /> | ||
| + | *'''Rrp44''' is the catalytic subunit <ref>PMID:19060898</ref>.<br /> | ||
| + | *'''Csl4''' is required for mRNA decay.<br /> | ||
| + | *'''M-phase phosphoprotein 6''' is involved in the 3'-processing of the 7S pre-RNA to the mature 5.8S rRNA. | ||
| + | |||
| + | __TOC__ | ||
== Function == | == Function == | ||
In order to see the structure of the protein complex, press <scene name='60/609824/2nn6/8'>Exosome</scene>. | In order to see the structure of the protein complex, press <scene name='60/609824/2nn6/8'>Exosome</scene>. | ||
| - | + | ===Enzymatic function=== | |
The exosome is primarily a 3'-5' exoribonuclease, meaning that it degrades RNA molecules from their 3' end. In eukaryotes it also have an endoribonucleolytic function, meaning it cleaves RNA at sites within the molecule. | The exosome is primarily a 3'-5' exoribonuclease, meaning that it degrades RNA molecules from their 3' end. In eukaryotes it also have an endoribonucleolytic function, meaning it cleaves RNA at sites within the molecule. | ||
| - | + | ===Substrates=== | |
The exosome is involved in the degradation and processing of a wide variety of RNA species. Substrates of the exosome include messenger RNA, ribosomal RNA, and many species of small RNAs. | The exosome is involved in the degradation and processing of a wide variety of RNA species. Substrates of the exosome include messenger RNA, ribosomal RNA, and many species of small RNAs. | ||
== Disease == | == Disease == | ||
| - | + | ===Autoimmunity=== | |
The exosome complex is the target of autoantibodies, which are known to be found in people that suffer from various autoimmune diseases (especially PM/Scl overlap syndrome). In the autoimune diseases, antibodies are mainly directed against two of the proteins of the complex, called PM/Scl-100 and PM/Scl-75. | The exosome complex is the target of autoantibodies, which are known to be found in people that suffer from various autoimmune diseases (especially PM/Scl overlap syndrome). In the autoimune diseases, antibodies are mainly directed against two of the proteins of the complex, called PM/Scl-100 and PM/Scl-75. | ||
| - | + | ===Cancer treatment=== | |
The Exosome is found to be inhibited by a cancer chemotherapy drug, which called antimetabolite fluorouracil. This drug is one of the most successful drugs for treating solid tumors. | The Exosome is found to be inhibited by a cancer chemotherapy drug, which called antimetabolite fluorouracil. This drug is one of the most successful drugs for treating solid tumors. | ||
| - | + | ===Neurological disorders=== | |
Mutations in Exosome component 3 cause pontocerebellar hypoplasia and spinal motor neuron disease. | Mutations in Exosome component 3 cause pontocerebellar hypoplasia and spinal motor neuron disease. | ||
| Line 28: | Line 38: | ||
The core of the Exosome complex is made of a ring, which is consisting of six RNases proteins (Rnase PH-like proteins), and other proteins are attached. | The core of the Exosome complex is made of a ring, which is consisting of six RNases proteins (Rnase PH-like proteins), and other proteins are attached. | ||
You can also view the complex in a <scene name="/12/3456/Sample/2">ribbon view</scene>. | You can also view the complex in a <scene name="/12/3456/Sample/2">ribbon view</scene>. | ||
| + | |||
| + | ==3D structures of exosome== | ||
| + | [[Exosome 3D structures]] | ||
</StructureSection> | </StructureSection> | ||
| + | |||
== References == | == References == | ||
<references/> | <references/> | ||
| + | [[Category: Topic Page]] | ||
Current revision
Exosome
| |||||||||||
References
- ↑ http://en.wikipedia.org/wiki/Exosome_complex#Structure
- ↑ Falk S, Tants JN, Basquin J, Thoms M, Hurt E, Sattler M, Conti E. Structural insights into the interaction of the nuclear exosome helicase Mtr4 with the pre-ribosomal protein Nop53. RNA. 2017 Sep 7. pii: rna.062901.117. doi: 10.1261/rna.062901.117. PMID:28883156 doi:http://dx.doi.org/10.1261/rna.062901.117
- ↑ Wang C, Liu Y, DeMario SM, Mandric I, Gonzalez-Figueroa C, Chanfreau GF. Rrp6 Moonlights in an RNA Exosome-Independent Manner to Promote Cell Survival and Gene Expression during Stress. Cell Rep. 2020 Jun 9;31(10):107754. PMID:32521279 doi:10.1016/j.celrep.2020.107754
- ↑ Schaeffer D, Tsanova B, Barbas A, Reis FP, Dastidar EG, Sanchez-Rotunno M, Arraiano CM, van Hoof A. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat Struct Mol Biol. 2009 Jan;16(1):56-62. doi: 10.1038/nsmb.1528. Epub 2008 Dec , 7. PMID:19060898 doi:http://dx.doi.org/10.1038/nsmb.1528