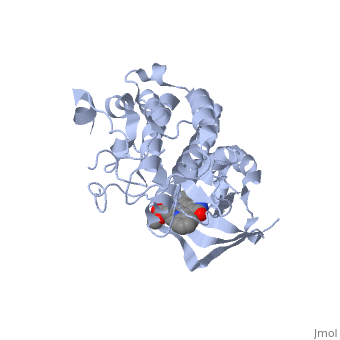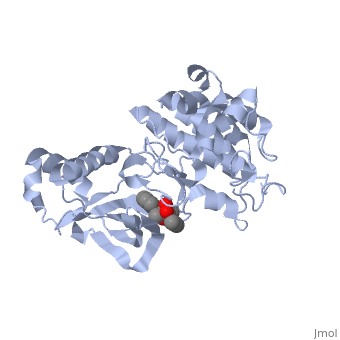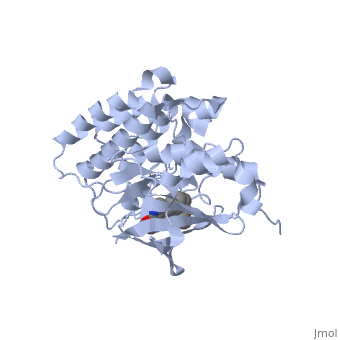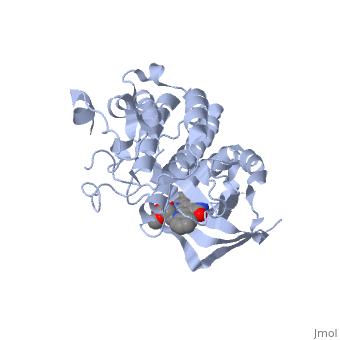
We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Hepatocyte growth factor receptor
From Proteopedia
(Difference between revisions)
| (9 intermediate revisions not shown.) | |||
| Line 3: | Line 3: | ||
==Introduction== | ==Introduction== | ||
| - | The '''Hepatocyte Growth Factor Receptor''' plays a large role in embryonic development, as its activation leads to events such as cell growth, motility and invasion. This receptor is a Tyrosine Kinase, and is one of the most well studied RTKs, as mutations in the c-met proto-oncogene can lead to the formation of tumors. The ligand for this receptor is Hepatocyte growth factor/scatter factor (HGF/SF), and upon binding of this ligand, the receptor becomes auto phosphorylated, causing downstream signaling events such as cell growth. <ref>PMID: 14559966</ref> C-Met is an αβ heterodimer with extracellular and intracellular domains. These two domains are disulfide linked together. <ref>PMID: 14559966</ref> This particular structure is one of a mutated hepatocyte growth factor tyrosine kinase domain, which is part of the intracellular β subunit. <ref>http://en.wikipedia.org/wiki/C-Met</ref> | + | The '''Hepatocyte Growth Factor Receptor''' or '''tyrosine-protein kinase Met''' or '''c-Met''' plays a large role in embryonic development, as its activation leads to events such as cell growth, motility and invasion. This receptor is a Tyrosine Kinase, and is one of the most well studied [[Receptor tyrosine kinases|RTKs]], as mutations in the c-met proto-oncogene can lead to the formation of tumors. The ligand for this receptor is Hepatocyte growth factor/scatter factor (HGF/SF), and upon binding of this ligand, the receptor becomes auto phosphorylated, causing downstream signaling events such as cell growth. <ref>PMID: 14559966</ref> C-Met is an αβ heterodimer with extracellular and intracellular domains. These two domains are disulfide linked together. <ref>PMID: 14559966</ref> This particular structure is one of a mutated hepatocyte growth factor tyrosine kinase domain, which is part of the intracellular β subunit.<ref>http://en.wikipedia.org/wiki/C-Met</ref>. See also [[Growth factors]] and [[Kinase-linked, enzyme-linked and related receptors]]. |
==Structure== | ==Structure== | ||
| Line 11: | Line 11: | ||
This receptor follows the typical structure of a protein kinase, with a bilobal structure. The N-terminal contains <scene name='Hepatocyte_growth_factor_receptor/Beta_sheets/1'>β-sheets</scene> and is linked through a hinge to the C lobe, which is full of α helices. This particular kinase domain is very similar to the domains of the insulin receptor kinase and fibroblast growth factor receptor kinase.<ref>PMID: 14559966</ref> | This receptor follows the typical structure of a protein kinase, with a bilobal structure. The N-terminal contains <scene name='Hepatocyte_growth_factor_receptor/Beta_sheets/1'>β-sheets</scene> and is linked through a hinge to the C lobe, which is full of α helices. This particular kinase domain is very similar to the domains of the insulin receptor kinase and fibroblast growth factor receptor kinase.<ref>PMID: 14559966</ref> | ||
| - | =Helices= | + | ==Helices== |
This structure is made up of many α helical structures that move in the transformation from inactive to active kinase. Some of these helices are conserved in many different tyrosine kinases. C-met does show a divergence from other tyrosine kinases (such as IRK and FGFRK) in the helix formed at the N-terminus, before the core kinase domain, in residues <scene name='Hepatocyte_growth_factor_receptor/1060-1069/1'>1060-1069</scene>. <ref>PMID: 14559966</ref> The αA is in contact with αC and so causes αC to be in a slightly different orientation than in FGFRK and IRK. Residues Leu-1062, Val-1066, and Val-1069 of αA <scene name='Hepatocyte_growth_factor_receptor/A_and_c_intercalating/1'>intercalate</scene> with with residues Leu-1125 and Ile-1129 of αC. There is another <scene name='Hepatocyte_growth_factor_receptor/A_and_c_intercalating/2'>interaction</scene> between the residues Ile-1053, Leu-1055 and Leu-1058 of αA and Ile-1118 and Val-1121 of αC. Because of the movement of αC during activation of the kinase, it is an assumption that αA is also part of the kinase activation upon ligand binding. <ref>PMID: 14559966</ref> | This structure is made up of many α helical structures that move in the transformation from inactive to active kinase. Some of these helices are conserved in many different tyrosine kinases. C-met does show a divergence from other tyrosine kinases (such as IRK and FGFRK) in the helix formed at the N-terminus, before the core kinase domain, in residues <scene name='Hepatocyte_growth_factor_receptor/1060-1069/1'>1060-1069</scene>. <ref>PMID: 14559966</ref> The αA is in contact with αC and so causes αC to be in a slightly different orientation than in FGFRK and IRK. Residues Leu-1062, Val-1066, and Val-1069 of αA <scene name='Hepatocyte_growth_factor_receptor/A_and_c_intercalating/1'>intercalate</scene> with with residues Leu-1125 and Ile-1129 of αC. There is another <scene name='Hepatocyte_growth_factor_receptor/A_and_c_intercalating/2'>interaction</scene> between the residues Ile-1053, Leu-1055 and Leu-1058 of αA and Ile-1118 and Val-1121 of αC. Because of the movement of αC during activation of the kinase, it is an assumption that αA is also part of the kinase activation upon ligand binding. <ref>PMID: 14559966</ref> | ||
| Line 19: | Line 19: | ||
<scene name='User:Juliette_Personius/sandbox_1/1234_and_1235_mutations/1'>tyr1234 and tyr1235</scene> are replaced by a phenylalanine and aspartate, respectively. This mutation normally causes the receptor to be constitutively active, and is found in HNSC (Head, Neck squamous cell) carcinoma. Although there is no longer phosphorylation at these sites, it is believed that the negative charge of the aspartate resembles the negative phosphate that would normally cause activation, and therefore keeps the protein in its active form. <ref>PMID: 14559966</ref> There is a third mutation at Tyr-1194 which is substituted for a <scene name='Hepatocyte_growth_factor_receptor/1194/1'>phenylalanine</scene>. This is shown to point into the <scene name='Hepatocyte_growth_factor_receptor/1198/1'>pocket</scene> formed by Lys-1198 and Leu-1195 from αE. <ref>PMID: 14559966</ref> This structure is conserved in the wild type protein, suggesting that the mutation at residue 1149 is not changing the structure at this position. | <scene name='User:Juliette_Personius/sandbox_1/1234_and_1235_mutations/1'>tyr1234 and tyr1235</scene> are replaced by a phenylalanine and aspartate, respectively. This mutation normally causes the receptor to be constitutively active, and is found in HNSC (Head, Neck squamous cell) carcinoma. Although there is no longer phosphorylation at these sites, it is believed that the negative charge of the aspartate resembles the negative phosphate that would normally cause activation, and therefore keeps the protein in its active form. <ref>PMID: 14559966</ref> There is a third mutation at Tyr-1194 which is substituted for a <scene name='Hepatocyte_growth_factor_receptor/1194/1'>phenylalanine</scene>. This is shown to point into the <scene name='Hepatocyte_growth_factor_receptor/1198/1'>pocket</scene> formed by Lys-1198 and Leu-1195 from αE. <ref>PMID: 14559966</ref> This structure is conserved in the wild type protein, suggesting that the mutation at residue 1149 is not changing the structure at this position. | ||
| - | =K-252a= | + | ==K-252a== |
<scene name='Hepatocyte_growth_factor_receptor/K-252a/1'>K-252a</scene> is a staurosporine analog. Staurosporine is an inhibitor of many Ser/Thr Kinases, and has been shown to also inhibit c-Met activation by inhibiting its autophosphorylation. The structures of K-252a and staurosporine are very similar, with the main difference being that K-252a has a furanose instead of a pyranose structure. The binding of K-252a causes the c-Met to adopt an inhibitory conformation of the A-loop, specifically with residues <scene name='Hepatocyte_growth_factor_receptor/Residues1231-1244/1'>1231-1244</scene> . This segment blocks the place where the substrate tyrosine side chain would bind, if the protein were in an active conformation. Residues | <scene name='Hepatocyte_growth_factor_receptor/K-252a/1'>K-252a</scene> is a staurosporine analog. Staurosporine is an inhibitor of many Ser/Thr Kinases, and has been shown to also inhibit c-Met activation by inhibiting its autophosphorylation. The structures of K-252a and staurosporine are very similar, with the main difference being that K-252a has a furanose instead of a pyranose structure. The binding of K-252a causes the c-Met to adopt an inhibitory conformation of the A-loop, specifically with residues <scene name='Hepatocyte_growth_factor_receptor/Residues1231-1244/1'>1231-1244</scene> . This segment blocks the place where the substrate tyrosine side chain would bind, if the protein were in an active conformation. Residues | ||
| Line 46: | Line 46: | ||
This structure also shows the binding motifs of c-Met in an unphosphorylated form, giving insight on how the motifs may move when interacting with their respective binding domains (Grb2, SH2, PTB domains). | This structure also shows the binding motifs of c-Met in an unphosphorylated form, giving insight on how the motifs may move when interacting with their respective binding domains (Grb2, SH2, PTB domains). | ||
| - | </StructureSection> | ||
== 3D Structures of hepatocyte growth factor receptor == | == 3D Structures of hepatocyte growth factor receptor == | ||
| + | [[Hepatocyte growth factor receptor 3D structures]] | ||
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | **[[1r0p]] – hHGFR tyrosine kinase domain (mutant) + K-252A <br /> | ||
| - | **[[2uzx]], [[2uzy]] – hHGFR SEMA, ψ, Ig1, Met741 domains + internalin B <br /> | ||
| - | **[[2rfn]], [[2rfs]], [[3ccn]], [[3cd8]], [[3f66]], [[2wd1]], [[2wgj]], [[3l8v]], [[3lq8]], [[2wkm]], [[3q6w]], [[3rhk]], [[3qti]], [[3zxz]], [[3zze]], [[3r7o]], [[3u6h]], [[3u6i]], [[4deg]], [[4deh]], [[4dei]], [[4aoi]], [[4ap7]], [[4gg5]], [[4gg7]], [[4eev]] – hHGFR tyrosine kinase domain + inhibitor <br /> | ||
| - | **[[3c1x]], [[3cth]], [[3ctj]], [[3ce3]], [[3efj]], [[3efk]], [[3f82]], [[3i5n]], [[3a4p]] – hHGFR tyrosine kinase domain (mutant) + inhibitor <br /> | ||
| - | **[[1shy]] – hHGFR SEMA and ψ domain + hHGF β chain – human <br /> | ||
| - | **[[3vw8]], [[3zbx]], [[3zc5]], [[3zcl]], [[4iwd]] - hHGFR tyrosine kinase domain + inhibitor<br /> | ||
| - | }} | ||
==References== | ==References== | ||
<references /> | <references /> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
References
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100
- ↑ http://en.wikipedia.org/wiki/C-Met
- ↑ Maina F, Casagranda F, Audero E, Simeone A, Comoglio PM, Klein R, Ponzetto C. Uncoupling of Grb2 from the Met receptor in vivo reveals complex roles in muscle development. Cell. 1996 Nov 1;87(3):531-42. PMID:8898205
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100