Molecular Playground/Human Serum Albumin
From Proteopedia
(Difference between revisions)
| (2 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | == '''HUMAN SERUM ALBUMIN (HSA)''' == | + | =='''HUMAN SERUM ALBUMIN (HSA)'''== |
| - | + | <StructureSection load='4K2C_all_domains_v2.pdb' size='500' side='right' caption='Human serum albumin (PDB code [[4iw1]])' scene=''> | |
| - | < | + | |
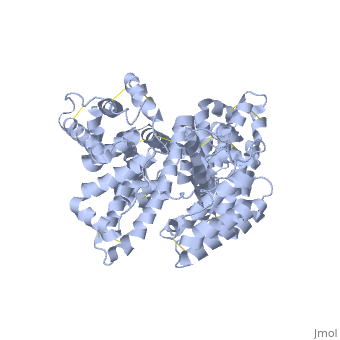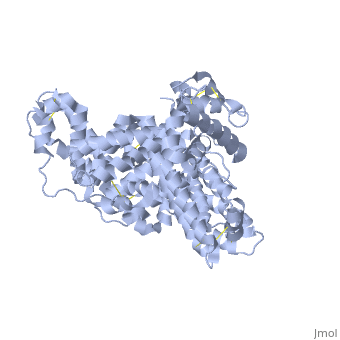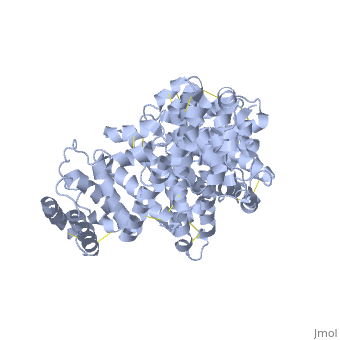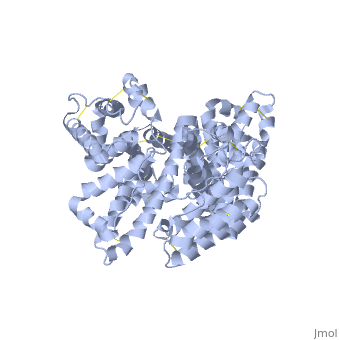
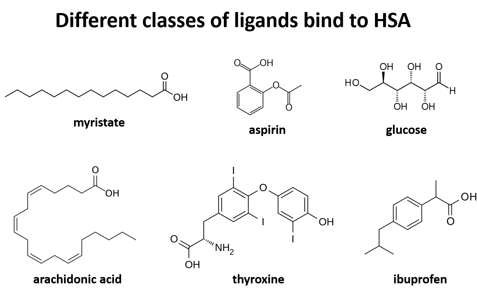
'''Human serum albumin''' (HSA) is the most abundant protein in the blood plasma, amounting to about 35 to 50 grams per liter of serum. With a molecular weight of about 66.5 kDa, it functions mainly to maintain the pH and osmotic pressure of the blood and to transport a wide variety of endogenous and exogenous substances. | '''Human serum albumin''' (HSA) is the most abundant protein in the blood plasma, amounting to about 35 to 50 grams per liter of serum. With a molecular weight of about 66.5 kDa, it functions mainly to maintain the pH and osmotic pressure of the blood and to transport a wide variety of endogenous and exogenous substances. | ||
| Line 9: | Line 8: | ||
HSA exists as a monomer that is comprised mostly of alpha helices. Each of the <scene name='57/571397/Different_domains_v2/1'>three homologous helical domains</scene> | HSA exists as a monomer that is comprised mostly of alpha helices. Each of the <scene name='57/571397/Different_domains_v2/1'>three homologous helical domains</scene> | ||
(<B><font color="purple">domain I</font></B>, <B><font color="orange">domain II</font></B>, <B><font color="cyan">domain III</font></B>) | (<B><font color="purple">domain I</font></B>, <B><font color="orange">domain II</font></B>, <B><font color="cyan">domain III</font></B>) | ||
| - | + | is further divided into subdomains A and B (A=darker shade, B=lighter shade), which then form several hydrophobic pockets throughout the molecule. This multidomain structure of HSA allows it to bind many different classes of ligands at multiple sites. | |
== Function == | == Function == | ||
| Line 24: | Line 23: | ||
== Removal of HSA in serum for biomarker studies == | == Removal of HSA in serum for biomarker studies == | ||
| - | Because HSA is very abundant in blood serum, most biomarker studies involving serum requires preliminary removal of HSA so that other non-abundant but otherwise important disease-indicating proteins can be detected. In our group, we exploit several properties of HSA in order to efficiently remove it from serum prior to biomarker analysis. These properties include its low isoelectric point (pI of about 4.7) due to a number of <scene name='57/571397/ | + | Because HSA is very abundant in blood serum, most biomarker studies involving serum requires preliminary removal of HSA so that other non-abundant but otherwise important disease-indicating proteins can be detected. In our group, we exploit several properties of HSA in order to efficiently remove it from serum prior to biomarker analysis. These properties include its low isoelectric point (pI of about 4.7) due to a number of <scene name='57/571397/Acidic_residues/1'>acidic residues</scene>, and its tendency to bind hydrophobic molecules through its hydrophobic pockets. |
Current revision
HUMAN SERUM ALBUMIN (HSA)
| |||||||||||