Porphobilinogen synthase
From Proteopedia
(Difference between revisions)
| (10 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
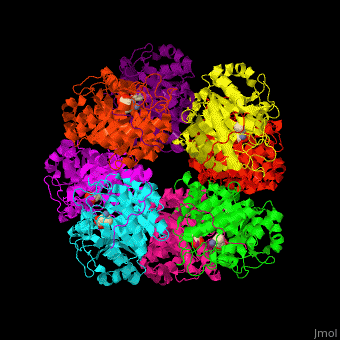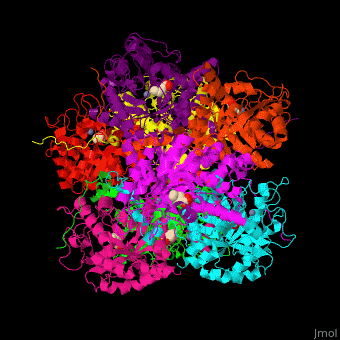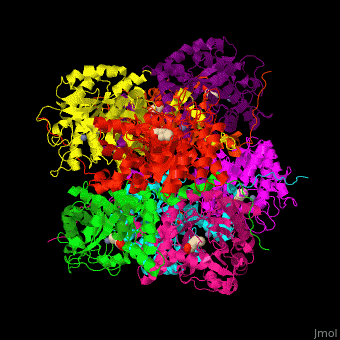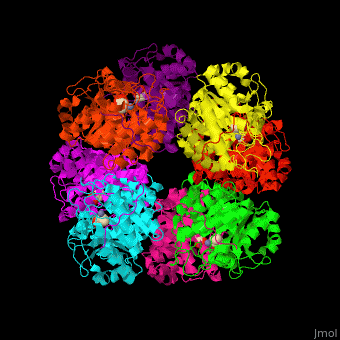
| - | + | <StructureSection load='' size='450' side='right' caption='Yeast porphobilinogen synthase complex with laevulunic acid and Zn+2 ion (grey) (PDB entry [[1h7n]])' scene='52/526179/Cv/1'> | |
| + | == Function == | ||
| + | '''Porphobilinogen synthase''' or '''δ-aminolaevulinic acid dehydratase''' (PBS) catalyzes the formation of porphobilinogen (PBG) from 2 molecules of aminolaevulinic acid (ALA). PBS participates in porphyrin biosynthesis<ref>PMID:15381398</ref>. Levulinic acid (LA) is an inhibitor of PBS. Porphyrin is the precursor of hemes, chlorophyll and vitamin B12. | ||
| - | + | == Disease == | |
| + | PBS deficiency can be caused by lead and other heavy metal poisoning<ref>PMID:7104498</ref>. | ||
| - | ===3D structures of porphobilinogen synthase | + | == Structural highlights == |
| + | The biological assembly of Yeast porphobilinogen synthase is <scene name='52/526179/Cv/4'>homooctamer</scene>. The <scene name='52/526179/Cv/5'>active site of PBS contains the substrate laevulinic acid and Zn+2 ion</scene><ref>PMID:11545591</ref>. Water molecules are shown as red spheres. | ||
| + | |||
| + | </StructureSection> | ||
| + | |||
| + | ==3D structures of porphobilinogen synthase== | ||
Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
{{#tree:id=OrganizedByTopic|openlevels=0| | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| Line 9: | Line 17: | ||
*Porphobilinogen synthase | *Porphobilinogen synthase | ||
| + | **[[5hms]], [[5hnr]] – hPBS + Zn – human<br /> | ||
| + | **[[1pv8]] – hPBS (mutant)<br /> | ||
| + | **[[2z0i]], [[2z1b]] - PBS - mouse<br /> | ||
**[[1aw5]] – yPBS (mutant) + Zn – yeast<br /> | **[[1aw5]] – yPBS (mutant) + Zn – yeast<br /> | ||
**[[1qml]] - yPBS + Hg<br /> | **[[1qml]] - yPBS + Hg<br /> | ||
**[[1qnv]] - yPBS + Pb<br /> | **[[1qnv]] - yPBS + Pb<br /> | ||
| - | + | **[[1w54]], [[1w56]], [[1w5m]], [[1w5n]], [[1w5o]], [[1w5p]], [[1w5q]] - PaPBS (mutant) + Mg + Zn – ''Pseudomonas aeruginosa''<br /> | |
| - | + | **[[5lzl]] - PBS + Zn – ''Pyrobaculum calidifontis''<br /> | |
| - | **[[1w54]], [[1w56]], [[1w5m]], [[1w5n]], [[1w5o]], [[1w5p]], [[1w5q]] - PaPBS (mutant) + Mg + Zn – ''Pseudomonas aeruginosa'' | + | |
*Porphobilinogen synthase complex | *Porphobilinogen synthase complex | ||
| - | **[[ | + | **[[1e51]] - hPBS + PBG + Zn<br /> |
| - | + | ||
**[[1ylv]], [[1h7n]] - yPBS + LA + Zn<br /> | **[[1ylv]], [[1h7n]] - yPBS + LA + Zn<br /> | ||
**[[1w31]] - yPBS + LA derivative + Zn<br /> | **[[1w31]] - yPBS + LA derivative + Zn<br /> | ||
| Line 25: | Line 34: | ||
**[[1h7r]], [[1h7p]], [[1eb3]], [[1gjp]] - yPBS + inhibitor + Zn<br /> | **[[1h7r]], [[1h7p]], [[1eb3]], [[1gjp]] - yPBS + inhibitor + Zn<br /> | ||
**[[1ohl]] - yPBS + intermediate + Zn<br /> | **[[1ohl]] - yPBS + intermediate + Zn<br /> | ||
| + | **[[1b4e]] - EcPBS + LA + Zn – ''Escherichia coli''<br /> | ||
| + | **[[5mhb]] - EcPBS + PBG + Zn<br /> | ||
**[[1i8j]], [[1l6s]], [[1l6y]] - EcPBS + inhibitor + Mg + Zn<br /> | **[[1i8j]], [[1l6s]], [[1l6y]] - EcPBS + inhibitor + Mg + Zn<br /> | ||
**[[2c1h]] - PBS + inhibitor + Mg – ''Chlorobium vibrioforme'' <br /> | **[[2c1h]] - PBS + inhibitor + Mg – ''Chlorobium vibrioforme'' <br /> | ||
**[[2c13]], [[2c14]] - PaPBS (mutant) + inhibitor + Mg<br /> | **[[2c13]], [[2c14]] - PaPBS (mutant) + inhibitor + Mg<br /> | ||
**[[2c15]], [[2c16]], [[2c18]], [[2c19]] - PaPBS + inhibitor + Mg<br /> | **[[2c15]], [[2c16]], [[2c18]], [[2c19]] - PaPBS + inhibitor + Mg<br /> | ||
| - | **[[ | + | **[[1b4k]] - PaPBS + LA + Zn<br /> |
**[[1gzg]] - PaPBS + LA derivative + Mg<br /> | **[[1gzg]] - PaPBS + LA derivative + Mg<br /> | ||
**[[2woq]] - PaPBS + antibiotic + Mg<br /> | **[[2woq]] - PaPBS + antibiotic + Mg<br /> | ||
| - | **[[1w1z]] - PBS + LA + Mg – ''Prosthecochloris vibrioformis'' | + | **[[1w1z]] - PBS + LA + Mg – ''Prosthecochloris vibrioformis''<br /> |
| + | **[[3obk]] - PBS residues 303-658 + Mg + PBG – ''Toxoplasma gondii''<br /> | ||
}} | }} | ||
| + | == References == | ||
| + | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
3D structures of porphobilinogen synthase
Updated on 06-October-2020
References
- ↑ Jaffe EK. The porphobilinogen synthase catalyzed reaction mechanism. Bioorg Chem. 2004 Oct;32(5):316-25. PMID:15381398 doi:http://dx.doi.org/10.1016/j.bioorg.2004.05.010
- ↑ Doss M, Muller WA. Acute lead poisoning in inherited porphobilinogen synthase (delta-aminolevulinic acid dehydrase) deficiency. Blut. 1982 Aug;45(2):131-9. PMID:7104498
- ↑ Erskine PT, Newbold R, Brindley AA, Wood SP, Shoolingin-Jordan PM, Warren MJ, Cooper JB. The x-ray structure of yeast 5-aminolaevulinic acid dehydratase complexed with substrate and three inhibitors. J Mol Biol. 2001 Sep 7;312(1):133-41. PMID:11545591 doi:10.1006/jmbi.2001.4947