Vascular Endothelial Growth Factor
From Proteopedia
(Difference between revisions)
| (33 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
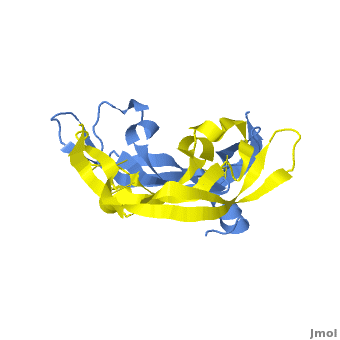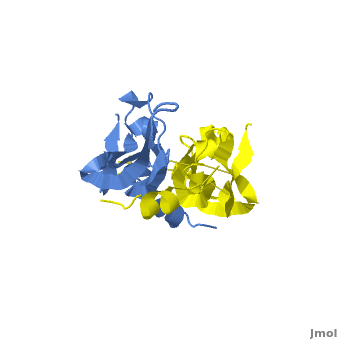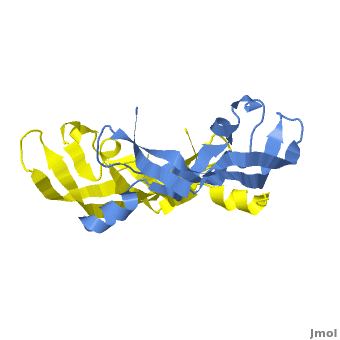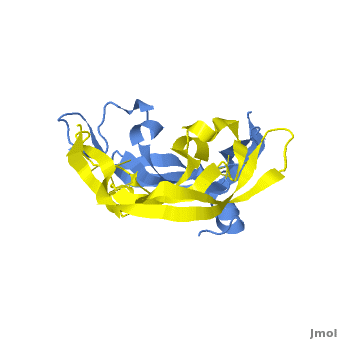
| - | <StructureSection load=' | + | <StructureSection load='.pdb' size='350' side='right' scene='Vascular_Endothelial_Growth_Factor/Vegf-a_opening/1' caption='Structure of Human VEGF-A dimer, [[1vpf]]'> |
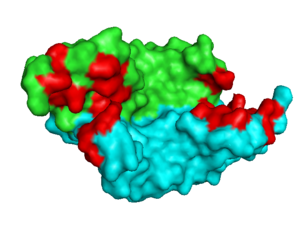
[[Image: VEGF_Opening.png|300px|left|thumb| VEGF-A highlighting 2 monomers and binding sites in red, [[1vpf]]]] | [[Image: VEGF_Opening.png|300px|left|thumb| VEGF-A highlighting 2 monomers and binding sites in red, [[1vpf]]]] | ||
| + | |||
{{Clear}} | {{Clear}} | ||
| - | [[Vascular Endothelial Growth Factor]]s (VEGFs) are a class of proteins that regulate vascular development in embryos and angiogenesis in adult mammals after sustaining an injury or notably in [[Cancer|cancerous]] tumors. A number of structural studies have been conducted on VEGF and its receptors (VEGFRs) to better understand the VEGF-VEGFR interaction and how the signal cascade originating from this interaction leads to a number of biological features. VEGF and its receptors have been closely looked at for their potential use as targets for pharmaceutical medicine with some success. The VEGF family contains VEGF-A which mediates increased vascular permeability | + | ==Introduction== |
| + | [[Vascular Endothelial Growth Factor]]s (VEGFs) are a class of proteins that regulate vascular development in embryos and angiogenesis in adult mammals after sustaining an injury or notably in [[Cancer|cancerous]] tumors. A number of structural studies have been conducted on VEGF and its receptors (VEGFRs) to better understand the VEGF-VEGFR interaction and how the signal cascade originating from this interaction leads to a number of biological features. VEGF and its receptors have been closely looked at for their potential use as targets for pharmaceutical medicine with some success. The VEGF family contains: | ||
| + | *'''VEGF-A''' which mediates increased vascular permeability<ref>PMID:33478167</ref>. | ||
| + | *'''VEGF-B''' which is a growth factor <ref>PMID:19684473</ref>. | ||
| + | *'''VEGF-C''' and '''VEGF-D''' are active in angiogenesis<ref>PMID:9708799</ref>,<ref>PMID:37686121</ref>. | ||
| + | *'''VEGF-E''' is found in viruses<ref>PMID:22507661</ref> | ||
| + | *'''VEGF-F''' found in snake venom<ref>PMID:15542594</ref>.<br /> | ||
| + | For additional information see:<br /> | ||
| + | [[VEGFR]]<br /> | ||
| + | [[VEGF IN COMPLEX WITH A NEUTRALIZING ANTIBODY]]<br /> | ||
| + | [[Autocrine signaling]]<br /> | ||
| + | [[VEGF signaling pathway]]. | ||
<br /> | <br /> | ||
| - | _TOC_ | ||
<br /> | <br /> | ||
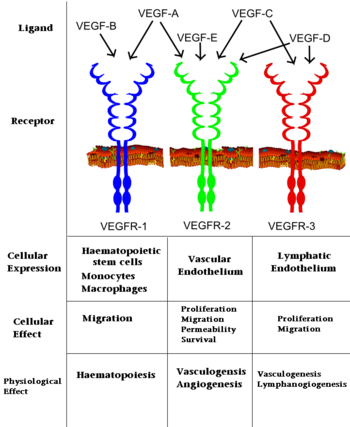
[[Image: VEGF_effects.PNG|350px|left|thumb| Interaction of VEGFs with VEGFRs. ]] | [[Image: VEGF_effects.PNG|350px|left|thumb| Interaction of VEGFs with VEGFRs. ]] | ||
| Line 25: | Line 36: | ||
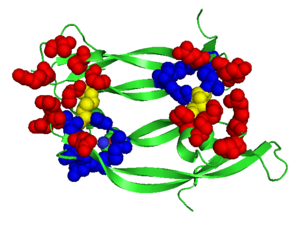
Although VEGF-E is only found in viral sources and thus is of less importance than VEGF-A, analysis of its structure is informative because it is the only member of the VEGF family that binds exclusively to VEGFR-2, the most essential VEGF receptor. Further, VEGF-E shares significant homology to VEGF-A, and thus can serve as an effective model. <ref>PMID:16672228</ref> | Although VEGF-E is only found in viral sources and thus is of less importance than VEGF-A, analysis of its structure is informative because it is the only member of the VEGF family that binds exclusively to VEGFR-2, the most essential VEGF receptor. Further, VEGF-E shares significant homology to VEGF-A, and thus can serve as an effective model. <ref>PMID:16672228</ref> | ||
| - | <scene name='Vascular_Endothelial_Growth_Factor/Vegf-e_opening/1'>VEGF-E</scene> consists of a homodimer that is covalently linked by two intermolecular disulfide bonds between <scene name=' | + | <scene name='Vascular_Endothelial_Growth_Factor/Vegf-e_opening/1'>VEGF-E</scene> consists of a homodimer that is covalently linked by two intermolecular disulfide bonds between <scene name='41/411433/Cys51-cys60/1'>Cys51 and Cys 60</scene>. |
| - | <scene name=' | + | |
| + | Each monomer contains a central antiparallel beta sheet, with the canonical <scene name='41/411433/Knot_new/3'>cysteine knot </scene> found in other VEGFs. <ref>PMID:1396586</ref> The knot consists of an eight residue ring formed by the backbone of residues 57-61 and 102-104 and intramolecular disulfide bridges Cys57-Cys102 and Cys61-Cys104, and a third bridge, Cys26-Cys68, that passes perpendicularly through the ring. Each VEGF-E monomer contains an amino terminal alpha helix and three solvent accessible loop regions, L2, <scene name='41/411433/Vegf-e_l1_l3/3'>L1 and L3 </scene>. | ||
| + | |||
| + | are able to form a complex hydrogen bond network as well as extensive hydrophobic contacts with VEGFR making these loops ideal receptor specificity determinants. Residues: P34, S36, T43, P50, R46, D63, E64, and E67 make up the <scene name='Vascular_Endothelial_Growth_Factor/Vegf-e_binding_site/1'>VEGF-E binding pocket </scene>and are critical for binding to VEGFR-2 as determined by alanine mutagenesis.<ref> PMID:16672228</ref> Further, the salt bridge between <scene name='41/411433/Vegf-e_salt_bridge/4'>R46 and E64 </scene> is believed to be the source of VEGF-E’s VEGFR-2 specificity by preventing binding to VEGFR-1. <ref>PMID:15272021</ref> | ||
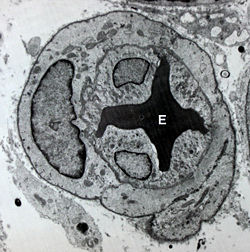
[[Image: Microvessel.jpg|250px|left|thumb| Image of a blood vessel with an erythrocyte (E) within its lumen and endothelial cells form its its tunica intima]] | [[Image: Microvessel.jpg|250px|left|thumb| Image of a blood vessel with an erythrocyte (E) within its lumen and endothelial cells form its its tunica intima]] | ||
| Line 34: | Line 48: | ||
VEGF has received significant attention from the pharmaceutical industry in the hopes of correcting impaired vessel function. Such impaired vessel function accompanies many pathologies including atherosclerosis, arthritis, some neurodegenerative diseases such as ALS, and malignant cell growth such as cancerous tumors. <ref> PMID:15488020</ref> In fact, numerous studies highlighted the increased expression of VEGF in cancer cells resulting in tumoral angiogenesis, providing the tumor with the network of blood vessels need to grow and expand. VEGF expression is stimulated by hypoxia, a common characteristic of most newly formed tumors, and genetic mutations such as [[K-ras]] and [[p53]], extremely common mutations present in a majority of cancers. <ref>PMID:20662002</ref> Since angiogenesis in typical adults is infrequent while extremely common in tumors, VEGF serves as a selective therapeutic target for cancer. A number of drugs, like Bevacizumab (a [[monoclonal antibody]] better known as [[Avastin]]) have been developed to interrupt the VEGF-VEGFR connection, with some success. Avastin binds to and inhibits all VEGF-A isoforms and has achieved megablockbuster status by earning over $5 billion in 2009 for Roche. <ref>http://www.foxbusiness.com/story/markets/industries/technology/roche-increased-avastin-sales-efforts-doubles-force/</ref> | VEGF has received significant attention from the pharmaceutical industry in the hopes of correcting impaired vessel function. Such impaired vessel function accompanies many pathologies including atherosclerosis, arthritis, some neurodegenerative diseases such as ALS, and malignant cell growth such as cancerous tumors. <ref> PMID:15488020</ref> In fact, numerous studies highlighted the increased expression of VEGF in cancer cells resulting in tumoral angiogenesis, providing the tumor with the network of blood vessels need to grow and expand. VEGF expression is stimulated by hypoxia, a common characteristic of most newly formed tumors, and genetic mutations such as [[K-ras]] and [[p53]], extremely common mutations present in a majority of cancers. <ref>PMID:20662002</ref> Since angiogenesis in typical adults is infrequent while extremely common in tumors, VEGF serves as a selective therapeutic target for cancer. A number of drugs, like Bevacizumab (a [[monoclonal antibody]] better known as [[Avastin]]) have been developed to interrupt the VEGF-VEGFR connection, with some success. Avastin binds to and inhibits all VEGF-A isoforms and has achieved megablockbuster status by earning over $5 billion in 2009 for Roche. <ref>http://www.foxbusiness.com/story/markets/industries/technology/roche-increased-avastin-sales-efforts-doubles-force/</ref> | ||
<br /> | <br /> | ||
| - | |||
| - | </StructureSection> | ||
| - | |||
== 3D Structures of VEGF== | == 3D Structures of VEGF== | ||
| + | [[VEGF 3D Structures]] | ||
| - | + | {{Clear}} | |
| - | + | ||
| - | *VEGF-A | ||
| - | |||
| - | **[[3bdy]], [[2qr0]] - hVEGF-A + hFab light+heavy chain – human<br /> | ||
| - | **[[4kzn]] - hVEGF-A receptor-binding domain<br /> | ||
| - | **[[2fjg]], [[2fjh]] - hVEGF-A receptor-binding domain+ hFab light+heavy chain <br /> | ||
| - | **[[3p9w]] - hVEGF-A + hFab heavy chain<br /> | ||
| - | **[[1tzh]], [[1tzi]], [[1cz8]] - hVEGF-A + mFab YADS1 light+heavy chain - mouse<br /> | ||
| - | **[[1mkg]], [[1mkk]], [[1mjv]] – hVEGF-A (mutant) <br /> | ||
| - | **[[3qtk]] - hVEGF-A residues 34-135<br /> | ||
| - | **[[4glu]] - hVEGF-A residues 1-102<br /> | ||
| - | **[[1kmx]] - hVEGF-A heparin-binding domain<br /> | ||
| - | **[[1vgh]], [[2vgh]] - hVEGF-A heparin-binding domain - NMR<br /> | ||
| - | **[[1qty]], [[1flt]] – hVEGF-A receptor-binding domain + hFMS-like tyrosine <br /> | ||
| - | **[[1vpp]] – hVEGF-A + receptor blocking peptide <br /> | ||
| - | **[[1bj1]] – hVEGF-A + mFab light+heavy chain fragments <br /> | ||
| - | **[[2vpf]], [[1vpf]] – hVEGF-A receptor binding domain<br /> | ||
| - | **[[1kat]] - hVEGF-A receptor binding domain + peptide antagonist - NMR<br /> | ||
| - | **[[3s1b]] - hVEGF-A + Mini-Z<br /> | ||
| - | **[[3s1k]] - hVEGF-A + Z-domain<br /> | ||
| - | **[[4gln]], [[4gls]] - hVEGF-A + D-RFX001<br /> | ||
| - | **[[4deq]] – hVEGF-A /neuropilin-1<br /> | ||
| - | |||
| - | *VEGF-B | ||
| - | |||
| - | **[[2xac]] – hVEGF-B receptor binding domain+ hVEGFR domain 2<br /> | ||
| - | **[[2vwe]] – hVEGF-B + mFab light+heavy chain fragments <br /> | ||
| - | **[[2c7w]] – hVEGF-B <br /> | ||
| - | **[[3v2a]] – hVEGF-B + VEGF-A<br /> | ||
| - | |||
| - | *VEGF-C | ||
| - | |||
| - | **[[2x1w]], [[2x1x]] – hVEGF-C VEGF homology domain+ VEGFR-2 domains 2 & 3<br /> | ||
| - | **[[4bsk]] - hVEGF-C VEGF homology domain + VEGFR-3 domains 1 & 2<br /> | ||
| - | |||
| - | *VEGF-D | ||
| - | |||
| - | **[[2xv7]] – hVEGF-D VEGF homology domain | ||
| - | |||
| - | *VEGF-E | ||
| - | |||
| - | **[[3v6b]] – VEGF-E + VEGFR 2 – Orf virus<br /> | ||
| - | |||
| - | *VEGF-F | ||
| - | |||
| - | **[[1wq8]], [[1wq9]] - VEGF-F – ''Vipera aspis'' | ||
| - | }} | ||
==Additional Resources== | ==Additional Resources== | ||
| - | For additional information, see: [[Cancer]] | + | For additional information, see: |
| - | + | * [[Cancer]] | |
| - | + | ||
| - | + | ||
* [[Vascular Endothelial Growth Factor Receptor]] | * [[Vascular Endothelial Growth Factor Receptor]] | ||
* [[Hormone]] | * [[Hormone]] | ||
| Line 98: | Line 61: | ||
==References== | ==References== | ||
<references /> | <references /> | ||
| - | + | </StructureSection> | |
| - | + | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, David Canner, Joel L. Sussman, Alexander Berchansky, Wayne Decatur, Jaime Prilusky