We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 959
From Proteopedia
(Difference between revisions)
| (113 intermediate revisions not shown.) | |||
| Line 2: | Line 2: | ||
{{Sandbox_Reserved_ESBS}} | {{Sandbox_Reserved_ESBS}} | ||
<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | <!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
| + | == <div class="center" style="width: auto; margin-left: auto; margin-right: auto;">Defensins-α-1</div> == | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | The calsequestrin 2 plays a major role here, because it regulates the release of the calcium in the cytosol while the membrane depolarization occurs and traps the calcium inside the lumen of the sarcoplasmic reticulum.<ref name="CASQ2 role">NCBI Gene Ressource: CASQ2 calsequestrin 2 http://www.ncbi.nlm.nih.gov/gene/845</ref> | ||
| - | It is also good to notice that a huge release of calcium in the cytosol would be lethal to the cell, since the calcium would precipitate with the free phosphate groups. | ||
| + | == Introduction == | ||
| + | Defensins (DEF) are a family of proteins which are involved in host defense in the epithelia of mucosal surfaces such as those of the intestin, respiratory tract, urinary tract, and vagina. They are antimicrobial and cytotoxic. All the proteins of the family are distinguished by a cystein motif and are encoded on the chromozome 8.<ref>http://www.ncbi.nlm.nih.gov/gene/1667</ref><br /> There are many defensins but in this article we will focus on the '''defensin-α-1'''. It is a polypeptide which is found in the microbicidal granules of neutrophils. It is synthesized in the neutrophils, which plays a role in the defense process. defensin-α-1 plays a particular role in phagocite-mediated host defense.<ref>''Abraham L. Kierszenbaum. Histologie et biologie cellulaire: Une introduction à l'anatomie pathologique'' 2002</ref> | ||
| - | == Structure == | ||
| - | === Monomere Structure === | ||
| - | Each monomer is divided in <scene name='56/568018/Monomer_structure/5'>3 thioredoxin domains (TRX)</scene>: <scene name='56/568018/Monomer_structure/9'>the N-term</scene>, <scene name='56/568018/Monomer_structure/10'>the middle</scene> and the <scene name='56/568018/Monomer_structure/11'>C-term</scene> domains. Each of these has a regular structure: a <scene name='56/568018/Beta_sheet/4'>5 strands beta sheet core</scene> surrounded by <scene name='56/568018/Alpha_helix/3'>4 alpha helices</scene>.<ref name="Martin">PMID:7788290</ref> | ||
| - | Usually these domains are involved in redox phenomena, which lead to disulfide bounds creation. Here these domains are inactive but play an important role in the polymerization of CASQ2.<ref name="Monomere structure">NCBI Structure Ressource: CASQ2 calsequestrin 2 http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?ascbin=8&maxaln=10&seltype=2&uid=239372&querygi=429544235&aln=1,227,0,109</ref> | ||
| - | Finally, the C-term Asp-rich end is intrisically disordered ''(therefore, the C-term end cannot be represented in 3D structures)''. <ref name="Polymerization of Calsequestrin: IMPLICATIONS FOR Ca2+ and REGULATION">Polymerization of Calsequestrin: IMPLICATIONS FOR Ca2+ and REGULATION (Park et al., 2003) http://www.jbc.org/content/278/18/16176.full.pdf+html</ref> | ||
| - | === Polymer Structure === | ||
| - | Within the sarcoplasmic reticulum (SR) lumen, CASQ2 polymerizes to form <scene name='56/568018/Dimer/1'>homodimers</scene>, homotetramers and | ||
| - | <scene name='56/568018/Oligomere_and_ligand/3'>homooligomers</scene>. | ||
| - | There are two forms of dimerization: the | ||
| - | <scene name='56/568018/Dimer/1'>front-to-front form</scene> and the <scene name='56/568018/Oligomere_and_ligand/5'>back-to-back form</scene>.<ref name="Crystal Structure of calsequestrin from rabbit skeletal muscle sarcoplasmic reticulum (Wang et al., 1998)">Crystal Structure of calsequestrin from rabbit skeletal muscle sarcoplasmic reticulum (Wang et al., 1998) http://www.nature.com/nsmb/journal/v5/n6/abs/nsb0698-476.html</ref> | ||
| - | The front-to-front one is stabilized by intermolecular interactions between the | ||
| - | <scene name='56/568018/Dimer/3'>α2 helix of the domain I</scene> of each CASQ2.<ref name="Crystal Structure of calsequestrin from rabbit skeletal muscle sarcoplasmic reticulum (Wang et al., 1998)">http://www.nature.com/nsmb/journal/v5/n6/abs/nsb0698-476.html</ref> The intermolecular salt bridges are built between <scene name='56/568018/Dimer/13'>Glu 55 and Lys 49</scene>.<ref name="Crystal Structure of calsequestrin from rabbit skeletal muscle sarcoplasmic reticulum (Wang et al., 1998)">http://www.nature.com/nsmb/journal/v5/n6/abs/nsb0698-476.html</ref> This dimerization induces the formation of an electronegative pocket which involves the following amino acids: Glu 39, Glu 54, Glu 78, Glu 92, Asp 93 and Asp 101 for the first monomere and Glu 199, Asp 245, Asp 278, Glu 348 and Glu 350 for the second one.<ref name="Crystal Structure of calsequestrin from rabbit skeletal muscle sarcoplasmic reticulum (Wang et al., 1998)">http://www.nature.com/nsmb/journal/v5/n6/abs/nsb0698-476.html</ref> | ||
| - | The back-to-back form is stabilized by intermolecular interactions between the <scene name='56/568018/Oligomere_and_ligand/7'>α3 helix of the domain I</scene>, <scene name='56/568018/Oligomere_and_ligand/6'>α4 helix of the domain II</scene><ref name="Crystal Structure of calsequestrin from rabbit skeletal muscle sarcoplasmic reticulum (Wang et al., 1998)">http://www.nature.com/nsmb/journal/v5/n6/abs/nsb0698-476.html</ref>, and it has also been proved that the <scene name='56/568018/Oligomere_and_ligand/18'>C-term domain</scene> is involved<ref name="c term">NCBI Structure Ressource: CASQ2 calsequestrin 2 http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi</ref> (<scene name='56/568018/Oligomere_and_ligand/9'>all together</scene>). The intermolecular salt bridges are built between Glu 215 and Lys 86, Glu 216 and Lys 24, Glu 169 and Lys 85.<ref name="Crystal Structure of calsequestrin from rabbit skeletal muscle sarcoplasmic reticulum (Wang et al., 1998)">http://www.nature.com/nsmb/journal/v5/n6/abs/nsb0698-476.html</ref> The dimerization is also favored by a hydrogen bond between Ala 82 and Asn 22. This dimerization creates a very electronegative pocket at the C-terminal region which enables the binding of Ca<sup>2+</sup>.<ref name="Crystal Structure of calsequestrin from rabbit skeletal muscle sarcoplasmic reticulum (Wang et al., 1998)">http://www.nature.com/nsmb/journal/v5/n6/abs/nsb0698-476.html</ref> | ||
| + | <StructureSection load='2pm4' size='450' side='right'caption='Crystal Structure of Defensin, (PDB code [[2pm4]]) '> | ||
| - | == | + | == Biological role == |
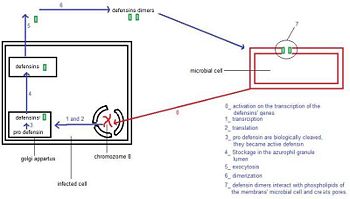
| + | The presence of an unknown microbial cell active the transcription of the defensin-α-1 gene in the human neutrophil. When the RNAm is translated defensins-α-1 are not active. They became active in the Golgi apparatus, in which they are biologically cleaved. Then they are stored in the azurophil granule lumen. After the exocytosis, two defensins-α-1 create a dimere, which will attack the membran of the microbial cell, by the formation of channel. These channels destabilize the membrane, which causes the destruction of the unknown microbial cell. | ||
| + | This mecanism is summed up in the following picture.<ref name="reactome">http://www.reactome.org/PathwayBrowser/#DIAGRAM=1461973&PATH=168256,168249</ref> | ||
| - | Each monomere of CASQ2 can bind between <scene name='56/568018/Oligomere_and_ligand/12'>18 to 50 Ca2+</scene>. The Ca<sup>2+</sup> ions bind to two or more acidic amino acids like <scene name='56/568018/Oligomere_and_ligand/13'>Glutamate</scene> or <scene name='56/568018/Oligomere_and_ligand/19'>Aspartate</scene>. These amino acids are mainly oriented outside and in the C-terminal region. It had been shown that Ca<sup>2+</sup>ions mainly bind an Asp-rich region on the disordered C-terminal domain. When CASQ2 form homooligomers, Ca<sup>2+</sup> can be bound in the electronegative pockets created by the <scene name='56/568018/Oligomere_and_ligand/17'>front-to-front</scene> and <scene name='56/568018/Oligomere_and_ligand/16'>back-to-back</scene> dimer interactions.<ref name="The asp-rich region at the carboxyl-terminus of calsequestrin binds to Ca2+ and interacts with triadin (Shin et al., 2000)">The Asp-rich region at the carboxyl-terminus of calsequestrin binds to Ca<sup>2+</sup> and interacts with triadin (Shin et al., 2000) http://www.sciencedirect.com/science/article/pii/S0014579300022468</ref> | ||
| - | + | {| align=center | |
| + | |- | ||
| + | | | ||
| + | [[Image: schema1.jpg|350px|left|thumb| General pathway of defensins-α-1.<ref name = "reactome"/>]] | ||
| + | {{clear}} | ||
| + | |} | ||
| - | == Interaction between CASQ2, Junctin and Triadin == | ||
| - | === | + | == Structure == |
| - | + | === Monomere structure === | |
| - | + | ||
| - | + | ||
| - | === | + | Defensin-α-1 are cationic antimicrobial peptides that are synthesized in vivo as inactive precursors. <br /> |
| - | + | Activation requires proteolytic excision of their anionic N-terminal inhibitory pro-peptide. The pro-peptide also specifically interacts with and inhibits the antimicrobial activity of the defensin-α-1 intermolecularly.<ref>www.ncbi.nlm.nih.gov/pmc/articles/PMC2754386/ )</ref> <br /> | |
| + | The active mature defensin-α-1 peptides consists of 29–35 amino acid residues with a molecular mass of 3–5 kDa.<br /> | ||
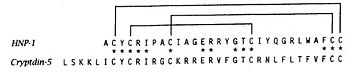
| + | The primary structure shows highly conserved residues, which are indispensable for the structural stability of the peptides. Among them are six invariant cysteine residues, necessary for the typical defensin-α-1 intramolecular disulphide-bond connectivity :<br /> | ||
| + | |||
| + | <div class="center" style="width: auto; margin-left: auto; margin-right: auto;"><scene name='60/604478/Cysteine2-30/1'>Cys2–Cys30</scene><br /></div> | ||
| + | <div class="center" style="width: auto; margin-left: auto; margin-right: auto;"><scene name='60/604478/Cysteine4-19/1'>Cys4–Cys19</scene><br /></div> | ||
| + | <div class="center" style="width: auto; margin-left: auto; margin-right: auto;"><scene name='60/604478/Cysteine9-26/1'>Cys9-Cys29</scene><br /></div> | ||
| + | {| align=center | ||
| + | |- | ||
| + | | | ||
| + | [[Image: disulfure.jpg|350px|left|thumb| Defensin-Alpha-1 cysteine connectivities.<ref name="stephen">Stephen H Wile, William C Wimley and Michael E Selsted. Structure, function, and membrane integration of defensins. Current Opinion in Structural Biology 1995. University of California, Irvine, USA</ref>]] | ||
{{clear}} | {{clear}} | ||
| - | + | |} | |
| + | There are two charged amino acid residues, <scene name='60/604478/Argglu/1'>Arg5, and Glu13</scene>, forming a conserved salt bridge, and <scene name='60/604478/Gly17/1'>Gly17</scene>, which constitutes a signature structural motif which is essential for correct folding. <br /> | ||
| - | + | Each defensin monomer consists of three strands of antiparallel β-sheet incorporating 60% of the residues. Two β-turns and three disulfide bonds add further restrictions to the conformational freedom of the monomer.<ref>http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2049026/</ref> | |
| - | + | === Dimere structure === | |
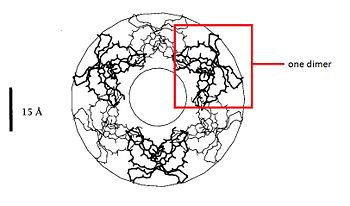
| + | The dimere is formed by joining identical β-strands of the two monomers together to create a symmetrical six-stranded β-sheet. This extended β-sheet twists and curls to form a basket-shaped structure that has a small solvent-accessible channel passing through it. The base of the basket is hydrophobic while the top, which contains the N- and C-terminal domains of the two defensin monomers, is polar. This dimer-of-dimers may be an essential feature of defensins’interaction with membranes.<ref name="gary">Gary Fujii, Michael E.Selsted, David Eisenberg. Defensins promote fusion and lysis of negatively charged membranes. Protein Science. 1993. Cambridge University</ref> | ||
| - | The | + | === Binding with the cell membrane === |
| + | The interaction of defensin-α-1 with cell membranes involves single dimer binding electrostatically to the cell surface. | ||
| + | In fact, the hydrophobic basket bottom of the defensin-α-1 dimer is inserted into the the hydrocarbon layer of one lipid monolayer while the polar group of the basket top and the <scene name='60/604478/Arg/1'>arginine</scene> maintain contact with the headgroup and aqueaus phase. Two dimers creat a channel. | ||
| + | The dimers can work together and create big channels like the following picture. The <scene name='60/604478/Hydrophobe/1'>hydrophobic residues</scene> are in the basket bottom.<ref name = "stephen"/> | ||
| + | {| align=center | ||
| + | |- | ||
| + | | | ||
| + | [[Image: membrane.jpg|350px|left|thumb|Creation of big channels..<ref name = "gary"/>]] ]] | ||
| + | {{clear}} | ||
| + | |} | ||
| + | |||
| + | == Example == | ||
| + | |||
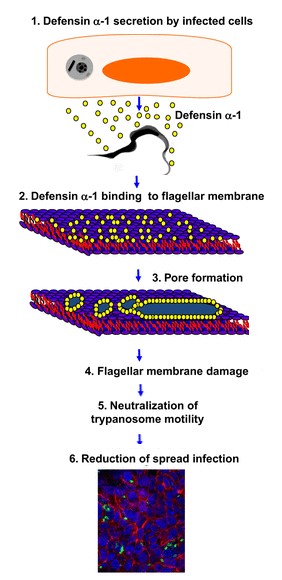
| + | DEF are known to play a role in the in the initiation of innate immune responses to some microbial pathogens. They are antimicrobial peptides of innate immunity functioning by non-specific binding to anionic phospholipids in bacterial membranes. For example defensins-α-1 have a role against the bacteria ''Trypanosoma cruzi''.<ref>http://iai.asm.org/content/81/11/4139.full/ref</ref><br /> | ||
| + | ''Trypanosoma cruzy'' or ''Cruzy'' debilitats Chagas disease, which affects millions of people and products significant morbidity and mortality. The defensins-α-1 are secreted by the HCT116 cells (which are Paneth cells), when they are infected by ''Cruzy''. They reduce the infection by making damage on the flagella structure. This damage inhibits parasite motility and reduces cellular infection. This reaction is introduced in the following drawing. | ||
| + | |||
| + | {| align=center | ||
| + | |- | ||
| + | | | ||
| + | [[Image: schema-defensins.jpg|350px|left|thumb| Defensins-α-1 pathway.]] | ||
| + | {{clear}} | ||
| + | |} | ||
| + | |||
| + | == Conclusion == | ||
| + | We focused on human defensin-α-1. It exists six different defensin-α. The first and the third differ only from one aminoacide. In addition of alpha defensins, there also exist Beta defensins which also play a role in defense. Defensins are extremely conserved so they are present by a great number of organisms. Researches in biotechnologies application with defensins are still performed in the fields of medecin and antibiotics. | ||
| - | </StructureSection> | ||
== References == | == References == | ||
<references /> | <references /> | ||
== '''Proteopedia page contributors and editors''' == | == '''Proteopedia page contributors and editors''' == | ||
| + | |||
| + | <div class="center" style="width: auto; margin-left: auto; margin-right: auto;">Julie Signoret and Stéphanie Gross - ChemBioTech</div> | ||
Current revision
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
Defensins-α-1
Introduction
Defensins (DEF) are a family of proteins which are involved in host defense in the epithelia of mucosal surfaces such as those of the intestin, respiratory tract, urinary tract, and vagina. They are antimicrobial and cytotoxic. All the proteins of the family are distinguished by a cystein motif and are encoded on the chromozome 8.[1]
There are many defensins but in this article we will focus on the defensin-α-1. It is a polypeptide which is found in the microbicidal granules of neutrophils. It is synthesized in the neutrophils, which plays a role in the defense process. defensin-α-1 plays a particular role in phagocite-mediated host defense.[2]
| |||||||||||