1yke
From Proteopedia
(Difference between revisions)
| (13 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | [[Image:1yke.gif|left|200px]] | ||
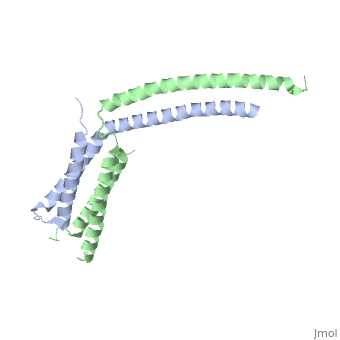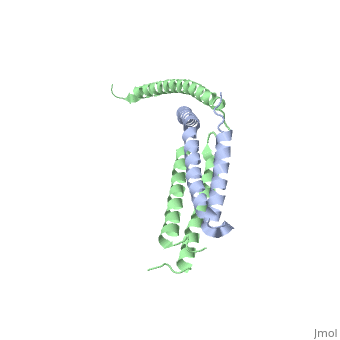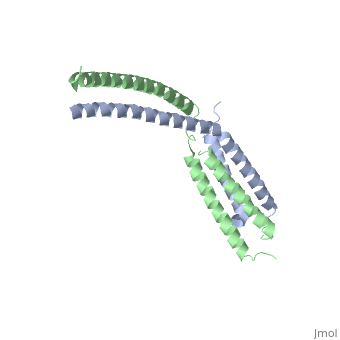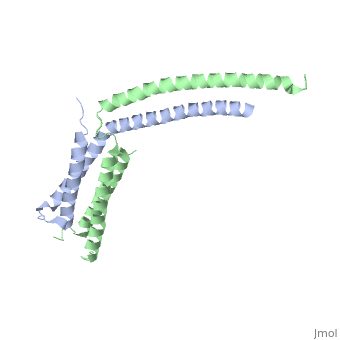
| - | + | ==Structure of the mediator MED7/MED21 subcomplex== | |
| - | + | <StructureSection load='1yke' size='340' side='right'caption='[[1yke]], [[Resolution|resolution]] 3.30Å' scene=''> | |
| - | + | == Structural highlights == | |
| - | + | <table><tr><td colspan='2'>[[1yke]] is a 4 chain structure with sequence from [https://en.wikipedia.org/wiki/Saccharomyces_cerevisiae Saccharomyces cerevisiae]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1YKE OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1YKE FirstGlance]. <br> | |
| - | | | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 3.3Å</td></tr> |
| - | | | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1yke FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1yke OCA], [https://pdbe.org/1yke PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1yke RCSB], [https://www.ebi.ac.uk/pdbsum/1yke PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1yke ProSAT]</span></td></tr> |
| - | + | </table> | |
| + | == Function == | ||
| + | [https://www.uniprot.org/uniprot/MED7_YEAST MED7_YEAST] Component of the Mediator complex, a coactivator involved in the regulated transcription of nearly all RNA polymerase II-dependent genes. Mediator functions as a bridge to convey information from gene-specific regulatory proteins to the basal RNA polymerase II transcription machinery. The Mediator complex, having a compact conformation in its free form, is recruited to promoters by direct interactions with regulatory proteins and serves for the assembly of a functional preinitiation complex with RNA polymerase II and the general transcription factors. The Mediator complex unfolds to an extended conformation and partially surrounds RNA polymerase II, specifically interacting with the unphosphorylated form of the C-terminal domain (CTD) of RNA polymerase II. The Mediator complex dissociates from the RNA polymerase II holoenzyme and stays at the promoter when transcriptional elongation begins.<ref>PMID:11555651</ref> <ref>PMID:16076843</ref> <ref>PMID:16263706</ref> | ||
| + | == Evolutionary Conservation == | ||
| + | [[Image:Consurf_key_small.gif|200px|right]] | ||
| + | Check<jmol> | ||
| + | <jmolCheckbox> | ||
| + | <scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/yk/1yke_consurf.spt"</scriptWhenChecked> | ||
| + | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | ||
| + | <text>to colour the structure by Evolutionary Conservation</text> | ||
| + | </jmolCheckbox> | ||
| + | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1yke ConSurf]. | ||
| + | <div style="clear:both"></div> | ||
| - | + | ==See Also== | |
| - | + | *[[Mediator|Mediator]] | |
| - | + | == References == | |
| - | == | + | <references/> |
| - | + | __TOC__ | |
| - | + | </StructureSection> | |
| - | == | + | [[Category: Large Structures]] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | [[Category: | + | |
[[Category: Saccharomyces cerevisiae]] | [[Category: Saccharomyces cerevisiae]] | ||
| - | [[Category: Baumli | + | [[Category: Baumli S]] |
| - | [[Category: Cramer | + | [[Category: Cramer P]] |
| - | [[Category: Hoeppner | + | [[Category: Hoeppner S]] |
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
Structure of the mediator MED7/MED21 subcomplex
| |||||||||||