We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
1fno
From Proteopedia
(Difference between revisions)
| (5 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| + | |||
==PEPTIDASE T (TRIPEPTIDASE)== | ==PEPTIDASE T (TRIPEPTIDASE)== | ||
| - | <StructureSection load='1fno' size='340' side='right' caption='[[1fno]], [[Resolution|resolution]] 2.40Å' scene=''> | + | <StructureSection load='1fno' size='340' side='right'caption='[[1fno]], [[Resolution|resolution]] 2.40Å' scene=''> |
== Structural highlights == | == Structural highlights == | ||
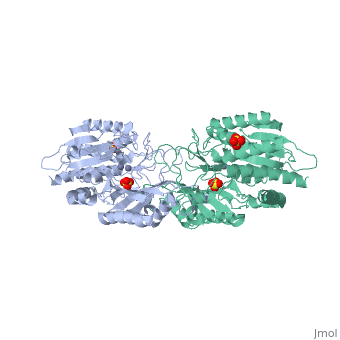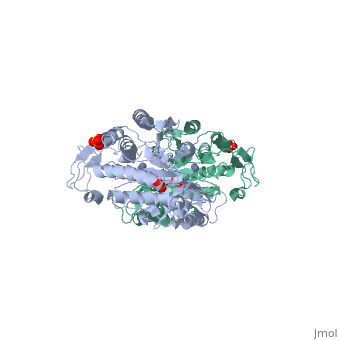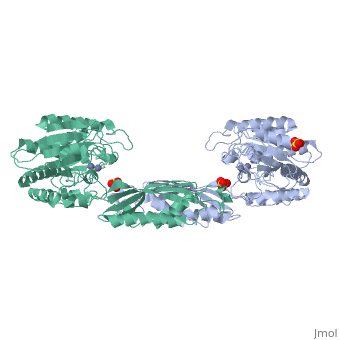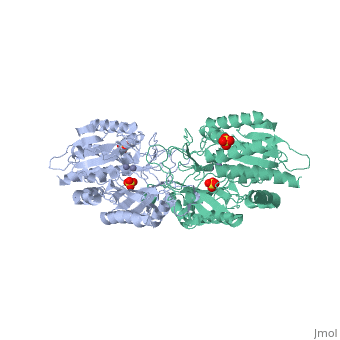
| - | <table><tr><td colspan='2'>[[1fno]] is a 1 chain structure with sequence from [ | + | <table><tr><td colspan='2'>[[1fno]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/Salmonella_enterica_subsp._enterica_serovar_Typhimurium Salmonella enterica subsp. enterica serovar Typhimurium]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1FNO OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1FNO FirstGlance]. <br> |
| - | </td></tr><tr id=' | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.4Å</td></tr> |
| - | <tr id=' | + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=MSE:SELENOMETHIONINE'>MSE</scene>, <scene name='pdbligand=SO4:SULFATE+ION'>SO4</scene>, <scene name='pdbligand=ZN:ZINC+ION'>ZN</scene></td></tr> |
| - | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1fno FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1fno OCA], [https://pdbe.org/1fno PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1fno RCSB], [https://www.ebi.ac.uk/pdbsum/1fno PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1fno ProSAT]</span></td></tr> |
</table> | </table> | ||
== Function == | == Function == | ||
| - | [ | + | [https://www.uniprot.org/uniprot/PEPT_SALTY PEPT_SALTY] Cleaves the N-terminal amino acid of tripeptides. Hydrolyzes tripeptides containing N-terminal methionine, leucine, or phenylalanine. Displays little or no activity against dipeptides, N-blocked or C-blocked tripeptides, and tetrapeptides.<ref>PMID:6341363</ref> |
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
Check<jmol> | Check<jmol> | ||
<jmolCheckbox> | <jmolCheckbox> | ||
| - | <scriptWhenChecked>select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/fn/1fno_consurf.spt"</scriptWhenChecked> | + | <scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/fn/1fno_consurf.spt"</scriptWhenChecked> |
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | ||
<text>to colour the structure by Evolutionary Conservation</text> | <text>to colour the structure by Evolutionary Conservation</text> | ||
</jmolCheckbox> | </jmolCheckbox> | ||
| - | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/ | + | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1fno ConSurf]. |
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
| - | <div style="background-color:#fffaf0;"> | ||
| - | == Publication Abstract from PubMed == | ||
| - | The structure of peptidase T, or tripeptidase, was determined by multiple wavelength anomalous dispersion (MAD) methodology and refined to 2.4 A resolution. Peptidase T comprises two domains; a catalytic domain with an active site containing two metal ions, and a smaller domain formed through a long insertion into the catalytic domain. The two metal ions, presumably zinc, are separated by 3.3 A, and are coordinated by five carboxylate and histidine ligands. The molecular surface of the active site is negatively charged. Peptidase T has the same basic fold as carboxypeptidase G2. When the structures of the two enzymes are superimposed, a number of homologous residues, not evident from the sequence alone, could be identified. Comparison of the active sites of peptidase T, carboxypeptidase G2, Aeromonas proteolytica aminopeptidase, carboxypeptidase A and leucine aminopeptidase reveals a common structural framework with interesting similarities and differences in the active sites and in the zinc coordination. A putative binding site for the C-terminal end of the tripeptide substrate was found at a peptidase T specific fingerprint sequence motif. | ||
| - | |||
| - | Structure of peptidase T from Salmonella typhimurium.,Hakansson K, Miller CG Eur J Biochem. 2002 Jan;269(2):443-50. PMID:11856302<ref>PMID:11856302</ref> | ||
| - | |||
| - | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
| - | </div> | ||
==See Also== | ==See Also== | ||
| Line 34: | Line 27: | ||
__TOC__ | __TOC__ | ||
</StructureSection> | </StructureSection> | ||
| - | [[Category: Salmonella enterica subsp. enterica serovar | + | [[Category: Large Structures]] |
| - | [[Category: Hakansson | + | [[Category: Salmonella enterica subsp. enterica serovar Typhimurium]] |
| - | [[Category: Miller | + | [[Category: Hakansson K]] |
| - | + | [[Category: Miller CG]] | |
| - | + | ||
| - | + | ||
Current revision
PEPTIDASE T (TRIPEPTIDASE)
| |||||||||||