We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
1e8o
From Proteopedia
(Difference between revisions)
| (5 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | == | + | |
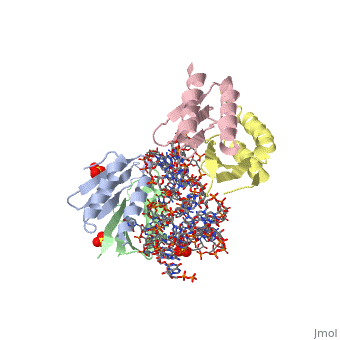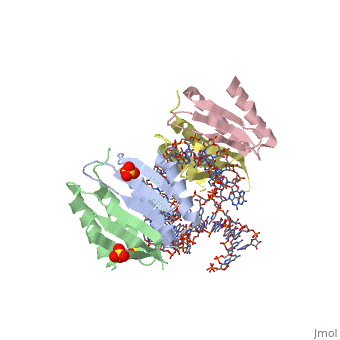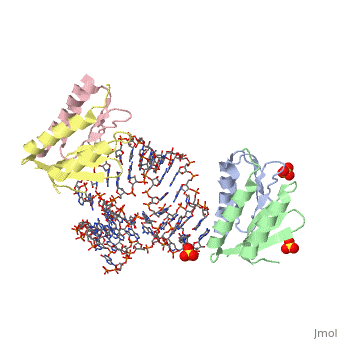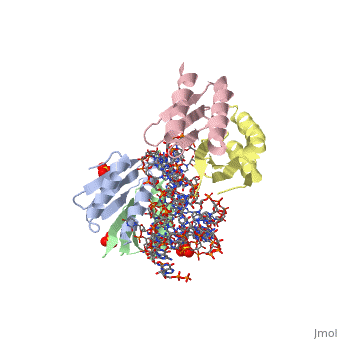
| - | <StructureSection load='1e8o' size='340' side='right' caption='[[1e8o]], [[Resolution|resolution]] 3.20Å' scene=''> | + | ==Core of the Alu domain of the mammalian SRP== |
| + | <StructureSection load='1e8o' size='340' side='right'caption='[[1e8o]], [[Resolution|resolution]] 3.20Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
| - | <table><tr><td colspan='2'>[[1e8o]] is a 5 chain structure with sequence from [ | + | <table><tr><td colspan='2'>[[1e8o]] is a 5 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1E8O OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1E8O FirstGlance]. <br> |
| - | </td></tr><tr id=' | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 3.2Å</td></tr> |
| - | <tr id=' | + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=GDP:GUANOSINE-5-DIPHOSPHATE'>GDP</scene>, <scene name='pdbligand=SO4:SULFATE+ION'>SO4</scene></td></tr> |
| - | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1e8o FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1e8o OCA], [https://pdbe.org/1e8o PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1e8o RCSB], [https://www.ebi.ac.uk/pdbsum/1e8o PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1e8o ProSAT]</span></td></tr> |
</table> | </table> | ||
== Function == | == Function == | ||
| - | [ | + | [https://www.uniprot.org/uniprot/SRP09_HUMAN SRP09_HUMAN] Signal-recognition-particle assembly has a crucial role in targeting secretory proteins to the rough endoplasmic reticulum membrane. SRP9 together with SRP14 and the Alu portion of the SRP RNA, constitutes the elongation arrest domain of SRP. The complex of SRP9 and SRP14 is required for SRP RNA binding. |
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
Check<jmol> | Check<jmol> | ||
<jmolCheckbox> | <jmolCheckbox> | ||
| - | <scriptWhenChecked>select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/e8/1e8o_consurf.spt"</scriptWhenChecked> | + | <scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/e8/1e8o_consurf.spt"</scriptWhenChecked> |
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | ||
<text>to colour the structure by Evolutionary Conservation</text> | <text>to colour the structure by Evolutionary Conservation</text> | ||
</jmolCheckbox> | </jmolCheckbox> | ||
| - | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/ | + | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1e8o ConSurf]. |
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
<div style="background-color:#fffaf0;"> | <div style="background-color:#fffaf0;"> | ||
| Line 27: | Line 28: | ||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
</div> | </div> | ||
| + | <div class="pdbe-citations 1e8o" style="background-color:#fffaf0;"></div> | ||
==See Also== | ==See Also== | ||
| + | *[[Signal recognition particle 3D structures|Signal recognition particle 3D structures]] | ||
*[[Signal recognition particle protein|Signal recognition particle protein]] | *[[Signal recognition particle protein|Signal recognition particle protein]] | ||
== References == | == References == | ||
| Line 35: | Line 38: | ||
</StructureSection> | </StructureSection> | ||
[[Category: Homo sapiens]] | [[Category: Homo sapiens]] | ||
| - | [[Category: Cusack | + | [[Category: Large Structures]] |
| - | [[Category: Strub | + | [[Category: Cusack S]] |
| - | [[Category: Weichenrieder | + | [[Category: Strub K]] |
| - | [[Category: Wild | + | [[Category: Weichenrieder O]] |
| - | + | [[Category: Wild K]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
Core of the Alu domain of the mammalian SRP
| |||||||||||