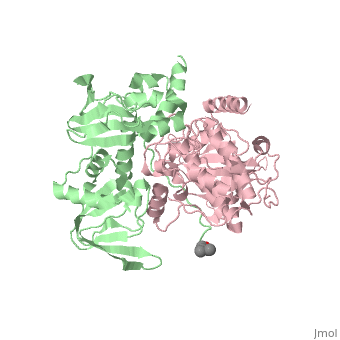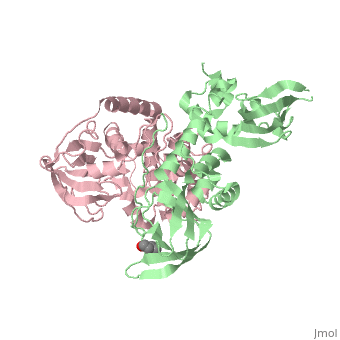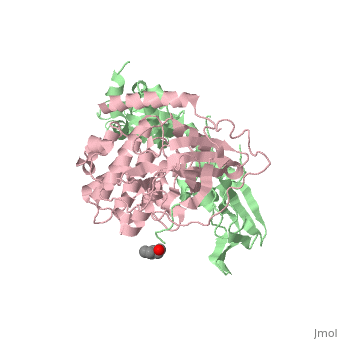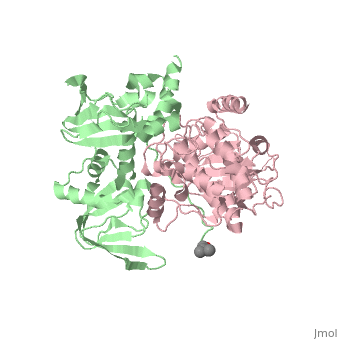Protein Kinase A
From Proteopedia
(Difference between revisions)
| (2 intermediate revisions not shown.) | |||
| Line 5: | Line 5: | ||
== Function == | == Function == | ||
Protein kinase A phosphorylates specific serine or threonine residues, which activates or deactivates the protein. Depending on the cell type, different proteins are available for phosphorylation. When activated with cAMP, PKA catalyzes the breakdown of glycogen, inhibits the synthesis of glucose, and promotes the increase of heart rate.In the "flight or fight" response, muscles become ready for action as a result of the activity of PKA. Overall, this enzyme helps regulate glucose levels,glycogen levels, sugar levels, and lipid metabolism. | Protein kinase A phosphorylates specific serine or threonine residues, which activates or deactivates the protein. Depending on the cell type, different proteins are available for phosphorylation. When activated with cAMP, PKA catalyzes the breakdown of glycogen, inhibits the synthesis of glucose, and promotes the increase of heart rate.In the "flight or fight" response, muscles become ready for action as a result of the activity of PKA. Overall, this enzyme helps regulate glucose levels,glycogen levels, sugar levels, and lipid metabolism. | ||
| + | See also [[CAMP-dependent pathway]] and [[CAMP is second messenger]]. | ||
== Structure == | == Structure == | ||
| Line 13: | Line 14: | ||
PKA is inactivated by a feedback mechanism. PKA activates phosphodiesterase, which converts cAMP back to ATP. This reduces the amount of cAMP that can activate PKA. | PKA is inactivated by a feedback mechanism. PKA activates phosphodiesterase, which converts cAMP back to ATP. This reduces the amount of cAMP that can activate PKA. | ||
| - | ==3D structures of protein kinase A | + | ==3D structures of protein kinase A== |
[[CAMP-dependent protein kinase]] | [[CAMP-dependent protein kinase]] | ||
Current revision
Protein kinase A (PKA)
| |||||||||||
References
[1]Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644 [2]C. Kim, N.-H. Xuong, and S.S. Taylor, "Crystal structure of a complex between catalytic and regulatory (Rlα) subunits of PKA,"Science 307, 690 (2005) doi:10.2210/rcsb_pdb/mom_2012_8