Sandbox Reserved 430
From Proteopedia
(Difference between revisions)
(→Binding Interactions) |
|||
| (109 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <!-- DO NOT DELETE THE TEMPLATE LINE --> | + | <references/><references/><!-- DO NOT DELETE THE TEMPLATE LINE --> |
{{Template:Sandbox Reserved Lynmarie Thompson}} | {{Template:Sandbox Reserved Lynmarie Thompson}} | ||
<!-- INSERT YOUR SCENES AND TEXT BELOW THIS LINE --> | <!-- INSERT YOUR SCENES AND TEXT BELOW THIS LINE --> | ||
| - | ==''' | + | =='''P2Y12 Receptor in Complex with AZD1283 (4ntj)<ref>PMID: 24670650 </ref>'''== |
| + | by [Cora Ricker, Lauren Timmins, Aidan Finnerty, Adam Murphy, Duy Nguyen] | ||
| + | |||
| + | [[Student Projects for UMass Chemistry 423 Spring 2016]] | ||
| + | <StructureSection load='4ntj' size='350' side='right' caption='Modeling of P2Y12 binded with Antithrombotic Drug([[4ntj]])' scene=''> | ||
==Introduction== | ==Introduction== | ||
| - | + | The goal of pharmaceuticals is to prevent or cure disease through drug therapy by specifically targeting cells, proteins, enzymes, genes, etc. It is often crucial to understand the structure, function, and relevant mechanisms involved with the target when designing an effective drug candidate. Furthermore, being able to know the effects on structure after drug-binding can provide insight into the functionality of a specific target. Because of this fact, it is common practice for research labs to develop protein crystals and use methods like x-ray diffraction, electron density mapping, and nuclear magnetic resonance spectroscopy to model the full structure. In this case, P2Y12, a member of P2Y receptor, was modeled when binded to AZD1283, an engineered receptor inhibitor.The molecular scene shows the chemical<scene name='48/483887/Here_is/1'> model</scene> of P2Y12 with the anionic side chains in red and charged nucleic acids and ligands in grey for contrast. | |
| - | + | Image analysis of P2Y12 crystals is used to model protein structure in complex with AstraZeneca’s novel AZD1283: Ethyl 6-(4(-((benzylsulphonyl)carbamoyl)piperidin-1yl)-5-cyano-2-methylnicotinate. AZD1283 functions to block the P2Y12 receptor as a means to treat thrombosis. AZD1283-binding leads to unique protein structure, unfound in other P2Y receptors. Helix V of seven transmembrane helices is found to be <scene name='48/483887/Helix_vii/1'>elongated and straightened</scene>. This change along with the discovery of a potential second active within P2Y12 has implications on how P2Y12 uses it’s seven transmembrane helical bundle interact with ADP in the bloodstream. | |
| - | + | As a member of a large class of G-protein-coupled receptors, P2Y12 is often an initial role player in signal transduction and cellular response due to external environmental factors. In the case of P2Y12, the receptor responds to ADP concentrations in the extracellular matrix and on a larger scale the blood stream. There is importance is understanding how P2Y12's structure receives ADP as an activator. In turn, knowledge of how P2Y12 is affected when properly inhibited can lead to improved drug design in terms of bioavailability, binding affinity, and effectiveness of inhibition. Ultimately, pharmaceuticals will be better able to prevent and treat cardiovascular diseases and medical conditions (thrombosis, hypercoagulable states) and more immediate dangers (stroke, embolism, and heart attacks). | |
| + | ==Overall Structure== | ||
| + | P2Y12R is a 1 chain structure. The <scene name='48/483887/Secondary_structure/2'>secondary structure</scene> of P2Y12R consists of eight <font color='deeppink'>alpha helices</font>. Seven transmembrane alpha helices are tilted and in a bundle, while the carboxy-terminal helix VIII is parallel to the membrane bilayer. The <scene name='48/483887/Rainbow/3'>rainbow scene</scene> demonstrates how the chain goes from the <font color='blue'>N</font> to <font color='red'>C</font> termini with each helix being approximately one color each of the color scheme. | ||
| + | {{Template:ColorKey_N2CRainbow}} | ||
| - | + | P2Y12R contains only one <scene name='48/483887/4ntj_disulfide_bond/1'>disulfide bond</scene> that connects the <font color='blue'>amino terminus</font> with <font color='darkorgange'>helix VII</font>. There are also two cholesterol molecules that are bound to two receptor molecules. As displayed in this <scene name='48/483887/Binding/2'>scene</scene>, one cholesterol molecule is bound to a receptor molecule between <font color='deepbluesky'>helix III</font> and <font color='lime'>helix V</font>. Another cholesterol molecule is bound to a receptor molecule shown <scene name='48/483887/Molecule/2'>here</scene> at the interface of <font color='blue'>helix I</font> and <font color='darkorange'>helix VII</font> . | |
| - | + | P2Y12R has some distinctive features from other GPCR structures in its family. <font color='lime'>Helix V</font>, for example, has around two more helical turns and does not have the typical helical bend that other GPCR structures have. As mentioned above, <font color='lime'>helix V</font> is <scene name='48/483887/Helix_vii/1'>elongated and straightened</scene> because the structure lacks proline and glycine residues to destabilize its structure. Furthermore, the elongated and straightened conformation causes P2Y12R’s extracellular end to shift 6 Å closer to <font color='turquoise'>helix IV</font> compared to other class A GPCR structures. In addition, the intracellular tip of <font color='darkorange'>helix VII</font> is closer to the seven transmembrane helical bundle. <font color='gold'>Helix VI’s</font> intracellular tip is tilted slightly outward and shifted closer to the intracellular surface than other GPCR structures. | |
| + | |||
| + | This <scene name='48/483887/Polar__nonpolar/1'>view</scene> demonstrates the polar and nonpolar regions of the P2Y12R's structure. AZD1283 spans more than 17 Å between <font color='turquoise'>helix IV</font> and <font color='darkorange'>helix VII</font> contributing to the polar and hydrophobic bonding with helices III–VI. | ||
| - | <br><br><br><br><br><br> | ||
| - | == | + | ==Binding Interactions== |
| - | < | + | The interaction between P2Y12 and AZD1283 is different in P2Y12 binding pocket and its PAR1 equivalent. PAR1's 24 residues of ECL2 have more interaction in <scene name='48/483887/Ligand/1'>ligand</scene>binding, while 16 unresolved residues ECL2 of P2Y12 is less likely to interact with AZD1283. Moreover, the shifted outward of helicies IV, VI and VII due to the extracellular make AZD1283 binds deeper into 7TM domain. It formed two pockets for the binding of AZD1283, separated by residues Y105 and K280, with pocket 1 consist of helices III-VII, while pocket 2 consists of helices I-III and VII. Among them, pocket 1 take part in the binding of AZD1283 and P2Y12, while pocket 2 does not. |
| - | + | The binding between P2Y12 and AZD1283 is also different from other GPCRs. The 17A elongated ligand is between helices IV and VII, which belongs in pocket 1. The antagonist AZD1283's piperidinyl-nicotinate group is inserted into sub-pocket of helices III, IV and V; while the benzylsulphonyl group interacts with helices VI and VII, forming at least seven polar and ionic interaction between P2Y12 and AZD1283. | |
| - | + | <scene name='48/483887/C97/1'>C97</scene> and C175 (two cysteine residues in helix III and ECL2 of P2Y12, respectively) are also notable in P2Y12-AZD1283 complexes. The two cysteines are highly conserved in GPCR family, and they form a disulphide bond in all GPCRs. But for P2Y12, no electron density is observed at C97, which means the disulphide bond in P2Y12 would be different. Moreover, mutation at C97 and C175 does not change the protein yield and stability, while increasing the melting temperature when in complex in AZD1283, which indicates that the mutation in both cysteine does not alter P2Y12 binding ability with AZD1283. | |
| - | <b><font color='red'>Acidity and Polarity</font></b> | ||
| - | This molecule has a dipole moment between the acidic and basic, or negatively and positively charged portions of the molecule. <scene name='48/483887/Acid_base/2'>Here</scene>, the <font color='purple'>acidic/negative areas are purple</font>, the <font color='green'>basic/positive areas are green</font>, and the <font color='brown'>neutral areas are golden brown</font>. These basic and acidic areas of the molecule alternate, just like the polar and non polar segments of chain alternate. | ||
| - | + | ==Additional Features== | |
| - | + | Acute coronary syndrome, a condition in which there is sudden blockage of blood flow to the heart, is majorly caused by a disease known as atherothrombosis. This disease is characterized by blocked arteries due to thrombosis: formation of a clot within a blood vessel. The P2Y12 protein is an important platelet receptor inhibitor that can be combined with aspirin in the management of ACS. It regulates certain functions through purinergic signaling, which is a form of extracellular signaling mediated by purine nucleotides and nucleosides like ADP and ATP.[5] | |
| - | + | The P2Y12 receptor is mainly found on the surface of blood platelets, and functions as a regulator in platelet activation and blood clotting. The P2Y12 receptor is a G-protein coupled receptor, <scene name='48/483887/Ss/1'>a seven-transmembrane domain protein</scene> (characterized by <font color='pink'>seven alpha helices each</font>) linked to the cAMP-signaling pathway. It mediates platelet activation by decreasing intracellular cAMP levels through inhibition of an AC-mediated signaling pathway.[1] Acting as a chemoreceptor, P2Y12 utilizes ADP as an agonist, which initiates ADP-induced platelet aggregation. Clopidogrel in covalent binding complex with P2Y12 acts as an anti platelet agent, specifically as an ADP receptor inhibitor to decrease platelet aggregation and inhibit thrombus formation.[3] | |
| - | < | + | |
| - | + | ||
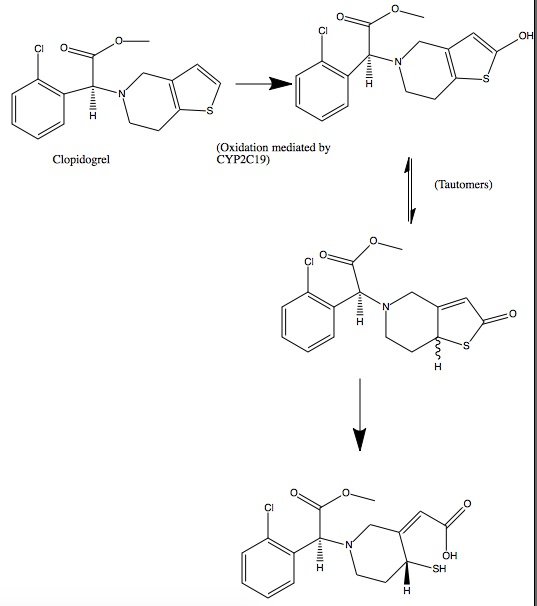
| - | + | Clopidogrel is a pro-drug and thienopyridine-type inhibitor of the P2Y12 receptor, which requires Cytochrome P450 to hepatically transform it to exert its anti platelet effect.[4] <scene name='48/483887/Aa/1'>Cytochrome P450</scene> is a membrane protein, characteristic of alternating <font color='gray'>hydrophobic</font> and <font color='pink'>polar</font> groups. The central heme group is stabilized by several side chains. The <font color='orange'>Fe2+</font> atom that makes up the heart of the heme group is surrounded by a <font color='blue'>highly hydrophobic porphyrin ring</font>. The role of the heme group in biological systems is to facilitate oxygen transport as well as aid in electron transfer as part of the electron transport chain. The heme group of the hemeprotein Cytochrome P450 acts as a catalyst for the metabolism of clopidogrel.[2] Shown below is the activation of Clopidogrel in vivo. CYP2C19 is an enzymatic member of the cytochrome P450 mixed-function oxidase system. The final step is a hydrolysis to yield the active metabolite. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | [[Image:Clop converted.jpg]] | |
| - | + | ||
| - | + | ||
| - | All reported small molecule Pim-1 inhibitors are known to competitive with ATP to bind with Pim-1 kinase. This competitive binding can occur via two different methods. One method is by closely mimicking the ATP binding to the hinge region of the kinase. Another method is using other areas of the active site to bind with the kinase <ref> Merkel, A. L.; Meggers, E.; Ocker, M. PIM1 kinase as a target for cancer therapy. Expert Opin. Investig. Drugs 2012, 21, 425-436.</ref>. The latter method still prevents ATP from binding with Pim-1.<br> | ||
| - | [[Image:ATP_and_Pim1_binding.jpg]]<br> | ||
| - | As shown in figure, ATP binds with the hinge region of the kinase. This hinge region lacks hydrogen bond donor and, therfore, can easily form hydrogen bond with ATP. <ref>Alex N. Bullock, Judit Debreczeni, Ann L. Amos, Stefan Knapp, and Benjamin E. Turk Structure and Substrate Specificity of the Pim-1 Kinase J. Biol. Chem. 2005 280: 41675-41682. First Published on October 13, 2005, doi:10.1074/jbc.M510711200</ref>. Primary amine group of ATP forms a hydrogen bond with the carbonyl oxygen of Glu 121. Knowing this simple interaction between ATP and Pim-1 kinase, scientists studied many small molecules that target this interaction. Aminooxadiazole-indole, the inhibitor of interest, was also designed to target this specific interaction as shown in this <scene name='48/483887/Hydrogen_bond/1'>scene</scene>. Aminooxadiazole not only forms a stable hydrogen bond with Glu 121, but it also interacts with other side chains around <scene name='48/483887/Active_site_4ty1/1'>active site</scene> of Pim-1. Binding to the kinase at the hinge region and other active sites, aminooxadiazole-indole inhibitor outcompetes ATP and effectively inactivates Pim-1. | ||
| - | |||
| - | <br> | ||
==Quiz Question 1== | ==Quiz Question 1== | ||
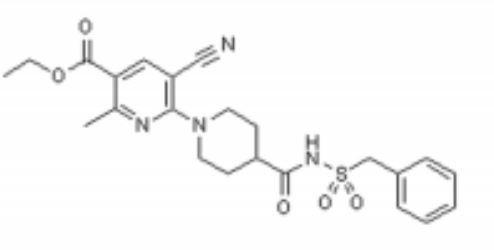
| - | < | + | Below is the structure for AZD1283. <scene name='48/483887/4ntj_azd_surroundingaas/5'>This scene </scene> shows a representation of the pocket where AZD1283 binds to P2Y12R. The dark salmon molecule is AZD1283 which is surrounded by magenta colored amino acids which are polar and gray colored ones which are hydrophobic.List 3 amino acid residues from the scene that should participate in hydrogen bonding with AZD1283 and 3 that should not. Why might the amino acids that don't hydrogen bond with AZD1283 still be present in the pocket? |
| - | < | + | |
| - | + | [[Image:AZD Structure.jpg]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==See Also== | ==See Also== | ||
| - | *[[ | + | *[[4ntj]] |
| - | *[[ | + | *[[4pxz]] |
| - | *[[ | + | *[[4py0]] |
| - | *[[ | + | *[[1vz1]] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==Credits== | ==Credits== | ||
| - | Introduction - | + | Introduction - Adam Murphy |
| - | Overall Structure - | + | Overall Structure - Cora Ricker |
| - | Drug Binding Site - | + | Drug Binding Site - Duy Nguyen |
| - | Additional Features - | + | Additional Features - Lauren Timmins |
| - | Quiz Question 1 - | + | Quiz Question 1 - Aidan Finnerty |
| - | + | ||
| - | + | ||
==References== | ==References== | ||
<references/> | <references/> | ||
| + | 1. Savi, P., et al. "P2Y 12, a new platelet ADP receptor, target of clopidogrel." Biochemical and biophysical research communications 283.2 (2001): 379-383. | ||
| + | |||
| + | 2. Coon, Minor J. "Cytochrome P450: nature's most versatile biological catalyst." Annu. Rev. Pharmacol. Toxicol. 45 (2005): 1-25. | ||
| + | |||
| + | 3. Gurbel, Paul A., et al. "Clopidogrel for coronary stenting response variability, drug resistance, and the effect of pretreatment platelet reactivity." Circulation 107.23 (2003): 2908-2913. | ||
| + | |||
| + | 4. Barn, Kulpreet, and Steven R. Steinhubl. "A brief review of the past and future of platelet P2Y12 antagonist." Coronary artery disease 23.6 (2012): 368-374. | ||
| + | |||
| + | 5. Bodin, Philippe, and Geoffrey Burnstock. "Purinergic signalling: ATP release." Neurochemical research 26.8-9 (2001): 959-969. | ||
Current revision
| This Sandbox is Reserved from January 19, 2016, through August 31, 2016 for use for Proteopedia Team Projects by the class Chemistry 423 Biochemistry for Chemists taught by Lynmarie K Thompson at University of Massachusetts Amherst, USA. This reservation includes Sandbox Reserved 425 through Sandbox Reserved 439. |
P2Y12 Receptor in Complex with AZD1283 (4ntj)[1]
by [Cora Ricker, Lauren Timmins, Aidan Finnerty, Adam Murphy, Duy Nguyen]
Student Projects for UMass Chemistry 423 Spring 2016
| |||||||||||