|
|
| (78 intermediate revisions not shown.) |
| Line 3: |
Line 3: |
| | <!-- INSERT YOUR SCENES AND TEXT BELOW THIS LINE --> | | <!-- INSERT YOUR SCENES AND TEXT BELOW THIS LINE --> |
| | | | |
| - | =='''Dengue virus methyltransferase bound to a SAM-based inhibitor-3P8Z'''== | + | =='''Vitamin D receptor/vitamin D (1db1)<ref>PMID: 10678179 </ref>'''== |
| | + | by Roger Crocker, Kate Daborowski, Patrick Murphy, Benjamin Rizkin and Aaron Thole |
| | + | |
| | + | |
| | + | [[Student Projects for UMass Chemistry 423 Spring 2016]] |
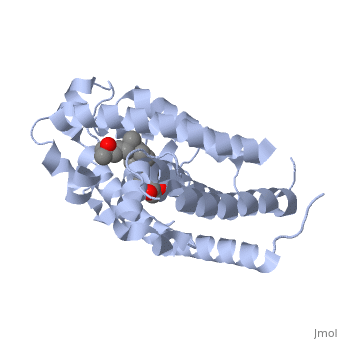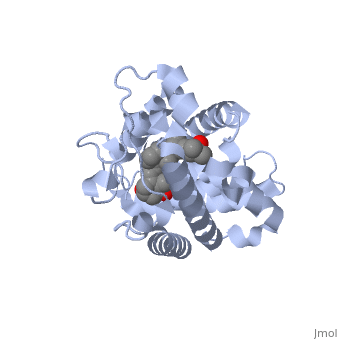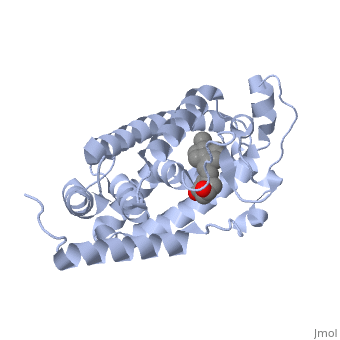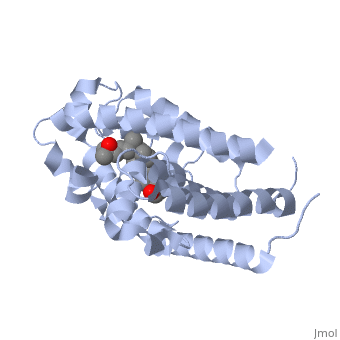
| | + | <StructureSection load='1db1' size='350' side='right' caption='caption for Molecular Playground (PDB entry [[1db1]])' scene=''> |
| | | | |
| | ==Introduction== | | ==Introduction== |
| | + | <br> |
| | + | The <scene name='48/483885/Color1/6'>vitamin D receptor</scene> (VDR) is a ligand-dependent transcriptional regulator with two strands. VDR belongs to the superfamily of nuclear receptors which control homeostasis, cell differentiation and growth, and many physiological processes. All proteins that belong to the nuclear receptor superfamily have a variable N-terminus region (A/B region), a hinge region that is flexible (D region), a conserved DNA-binding region (DBD, C region), and a moderately conserved ligand-binding region (LBD, E/F region). In the case of VDR, the A/B region is very short so it does not have any AF-1 function and the ligand binding region has a dimerization interface and a transcriptional activation domain that is ligand-dependent (AF-2).[1] <br> <br> |
| | + | The VDR has both an active and suppressed form. The activation or suppression function is caused by the binding of the DR3 response element as a heterodimer with the retinoid X receptor of the target genes. Due to the interactions with the basal transcriptional machinery and transcriptional cofactors, transcription is either activated or suppressed. When VDR is in its active form it regulates both phosphate and calcium metabolism, has immunosuppressive effects, and induces cell differentiation. When there are defects in the VDR that effect its metabolism it can lead to diseases such as severe rickets, secondary hyperparathyroidism, and hypocalcemia. Though defects in VDR can cause many diseases, fully functioning VDR can be used as treatment for disease such as cancer, autoimmune disease, psoriasis, osteoporosis, and renal osteodystrophy.[1] |
| | | | |
| - | Dengue virus, from the genus Flativirus, is a type of virus that causes yellow fever, dengue fever, tick-borne encephalitis, and Japanese B. encephalitis.<ref>AnonymousFlavivirus. http://www.britannica.com/EBchecked/topic/209809/flavivirus (accessed April 2, 2015). </ref> <Structure load='3p8z' size='300' frame='true' align='right' caption='3P8Z, Dengue virus Methyltransferase (MTase) bound to a SAM-based inhibitor' scene='48/483885/Initial_scene/1' /> | |
| - | <scene name='48/483885/Initial_scene/1'>Dengue virus Methyltransferase (MTase) bound to a SAM-based inhibitor</scene> is developed as an antiviral drug to target dengue virus infection by modulating flavivirus MTase. | |
| - | Dengue virus MTase is essential for viral replication because it has been found to catalyze two distinct methylation reactions at the guanine N7 and ribose 2’O position of the viral RNA cap structure. S-adenosyl-L-methionine (SAM) on the viral enzyme acts as a methyl group donor during methylation reaction due to its highly favorable reaction energetic.<ref>Schmidt, T.; Schwede, T.; Meuwly, M. Computational Analysis of Methyl Transfer Reactions in Dengue Virus Methyltransferase. J Phys Chem B 2014, 118, 5882-5890. DOI: 10.1021/jp5028564</ref> Dengue virus MTase bound to a SAM-based inhibitor is able to control the viral infection of dengue virus by preventing SAM from methylating and limiting the viral replication. This enzyme has a total of two chains that are represented in a 1-sequence-unique entity. It has two different types of ligand which are <scene name='48/483885/Initial_scene/3'>S-Adenosil-homocysteine (SAH) and (S)-2-amino-4-(((2S,3S,4R,5R)-5-(6-(3-chlorobenzylamino)-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methylthio)butanoic acid, (36A)</scene>. SAH known to be an inhibitor of the methylation reaction of the virus. Chemical derivatizations of SAH have been shown to have increased selective activity towards Dengue Virus Methyltransferase, while demonstrating no activity toward human enzymes. This reduces harmful or toxic activity in the human body.Since ligands 36A and SAH have similar chemical structure, it is predicted that ligands 36A and SAH each limits methylation at different position of the viral RNA. <ref>Lim SP, Sonntag LS, Noble C, Nilar SH, Ng RH, Zou G, Monaghan P, Chung KY, Dong H, Liu B, Bodenreider C, Lee G, Ding M, Chan WL, Wang G, Jian YL, Chao AT, Lescar J, Yin Z, Vedananda TR, Keller TH, Shi PY. Small molecule inhibitors that selectively block dengue virus methyltransferase. J Biol Chem. 2011 Feb 25;286(8):6233-40. Epub 2010 Dec 8. PMID:21147775 doi:10.1074/jbc.M110.179184</ref> | |
| | | | |
| - | | |
| - | <br><br><br><br><br><br><br><br><br><br><br> | |
| | | | |
| | ==Overall Structure== | | ==Overall Structure== |
| - | <Structure load='1a84' size='300' frame='true' align='right' caption='3p8z, Overall Structure' scene='48/483885/Polarity_3p8z/2' /> | + | The <scene name='48/483885/Alphabeta_picture/1'>vitamin d receptor</scene> contains 427 amino acids with a total molecular weight of 48,289 Da. The protein is also composed almost entirely of alpha helices with only a single beta sheet. The vitamin D receptor also does not have a quaternary structure [2]. <scene name='48/483885/Vitamin_d3/2'>Vitamin D3</scene> is a large organic compound made up of 27 carbon atoms, 44 hydrogen atoms and a single oxygen atom, with the ligand having a total molecular weight of 385 Da [3]. In studying the vitamin d receptor, the regions of the protein have been categorized into domains, with the A/B domain located at the N-terminus, the C domain, which is located between amino acid 20 and amino acid 115, the D domain, which is located between the end of the C domain and amino acid 220, and the EF domain, which encompasses the rest of the protein [4]. |
| - | Dengue Virus Methyltransferase bound to a SAM-Based inhibitor is a dimer, with two egg-like halves pointing at each other via their smaller point. The protein is composed of fourteen alpha helices, seven in each half of the dimer. There are eighteen beta sheets, nine in each half, in the protein. <scene name='48/483885/Polarity_3p8z/2'>The Green Scene</scene> illustrated below shows the charged groups in maroon and the hydrophobic groups are shown in lime green. The alpha helices are towards the outside of each half of the protein dimer and are both polar and non-polar. The beta sheets are closer to the insides of the two halves and are mostly non-polar. Almost all of the polar amino acids are on the outside of the protein, where the non-polar groups are both on the inside and outside of the protein.
| + | <br> <br> |
| - | | + | Other aspects of interest about the vitamin D receptor include the protein revealing a binding pocket when it is in its active folded state, allowing the ligand to bind to the receptor. The ligand interacts with the activation helix by stabilizing the agonist position. This is accomplished through Van der Waals interactions between the ligand and the activation helix. The activation ligand is a nuclear receptor. There is also some empty space observed around the aliphatic chain [1]. |
| - | | + | The protein has an active conformation of 1,25 (OH)_2D_3 that has a ligand binding pocket in its active folded state. The activation ligand, a nuclear receptor (VDR), interacts with with the activation helix by stabilizing the agonist position. This is accomplished through Van der Waals interactions between the ligand and the activation helix. There is some empty space observed around the aliphatic chain, indicating the presence of water to stabilize all possible hydrogen bonds [1]. |
| - | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | + | |
| | | | |
| | ==Binding Interactions== | | ==Binding Interactions== |
| - | | + | Protein 1db1 is found to complex with 1,25 Dihydroxy <scene name='48/483885/Vitamin_d3/1'>vitamin D3</scene>. This molecule has three notable alcohol groups shown in red. Oxygen is electronegative, giving alcohols the ability to participate in hydrogen bonding with the protein. Vitamin D3 has a large number of relations with the residues on the protein chain. First in the sequence are <scene name='48/483885/Residues_140-151/2'>residues 140-151</scene>. Tyr143, shown in blue, is the closest to the ligand at 2.83 angstroms. This is sightly large but there is still the possibility of hydrogen bonding. Tyr147 in green and Phe150 in black are also known to have interactions with vitamin D3 they are farther away and therefore less significant. Next down the peptide chain are <scene name='48/483885/Residues_235-240/1'>residues 235-240</scene>. Ser237, shown in green, has significant interactions with vitamin D3 this can be seen by its short distance 2.78 angstroms. Only 40 residues away, more hydrogen bonding is occurring.<scene name='48/483885/Residues_270-280/1'>Arg274 and Ser278</scene> form bonds with the same oxygen atoms as Tyr143 and Ser237 respectively. This means that the oxygen atoms of vitamin D3, when bound to the receptor, are negatively charged and stabilized by protons. On the opposite end of vitamin D3 there is an additional negatively charged oxygen. Although this oxygen does not participate in hydrogen bonding. <scene name='48/483885/His305_and_his397/1'>His305 and His397</scene>, shown in blue and green respectively, Contain aromatic rings. These rings are able to momentarily accept the electrons donated by the oxygen because they can delocalize the charge. This creates two pseudo-covalent bonds that is approximately 2.81 angstroms. When looking at <scene name='48/483885/All_binding_interactions/1'>all binding interactions</scene>, it can be seen that all of the binding sights are centered around oxygen [5,6]. |
| - | <Structure load='3p8z' size='300' frame='true' align='right' caption='3p8z, Binding Interaction' scene='48/483885/Protein_with_inhibitor/2' /> | + | |
| - | | + | |
| - | This viral protein functions to methylate the viral RNA cap of originating from the dengue virus. In order to do this, the protein binds a S-adenosyl-l-methionine ligand (SAM). The SAM ligand acts as the methyl group donor. It must also bind the 5' end of the RNA cap. MTases are highly conserved, thus blocking only the desired viral protein can prove to be difficult. | + | |
| - | | + | |
| - | In order to block this binding a similar ligand to the SAM ligand was employed. The ligand chosen as an inhibitor is the S-Adenosyl-L-homocysteine ligand. This ligand is similar to the SAM ligand with the one exception of a hydrogen where the methyl group donor is in the native substrate. The protein without the inhibitor bound is in <scene name='48/483885/Protein_without_inhibitor/1'>this </scene> conformation.<ref>http://proteopedia.org/wiki/index.php/4r8r</ref>. <ref>http://ac.els-cdn.com/S0166354214002563/1-s2.0-S0166354214002563-main.pdf?_tid=26b60158-c357-11e4-9d55-00000aab0f6b&acdnat=1425574160_e6994207ddfee772f88490227e693ef4</ref>
| + | |
| - | | + | |
| - | <scene name='48/483885/Protein_with_inhibitor/3'> Binding of the SAM inhibitor </scene> does not change the structure of the protein to a great degree, the RMS difference of the Cα atoms is 0.49 angstroms. | + | |
| - | | + | |
| - | The relatively <scene name='48/483885/3p8z_binding/3'>hydrophilic SAM ligand binds into the deep pocket</scene>. Favorable hydrogen bonds are satisfied in this interaction along with hydrophobic interactions between the non-polar residues and the non-polar portions of the inhibitor. The bonds are satisfied with the following amino acids on the protein, Phe-133, Ile-147, Gly-148, Glu-149, Arg-160, Arg-163, Val-164, and Leu-182. These residues help to stabilize the charges present on the ligand; they are not all strictly hydrophilic, however they do have portions that allow for hydrogen bonding.<ref>http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3057852/</ref>
| + | |
| - | | + | |
| - | Competitive binding on this active site could prove to be an effective drug target; if the SAM ligand cannot bind then the viral RNA cap will not be methylated. This could lead to a less viable virus due to a incorrectly formed 5' cap on the viral RNA.
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | <br><br><br><br><br><br>
| + | |
| | | | |
| | ==Additional Features== | | ==Additional Features== |
| | | | |
| - | <Structure load='3p8z' size='300' frame='true' align='right' caption='3p8z, Additional Features' scene='48/483885/Denv-3_mtase/2'/>
| + | Hereditary Vitamin D Resistant Rickets (HVDRR) is a condition that most commonly occurs in children and results in soft and weak bone formation, which often causes deformities in bone structure. A lack of proper nutrients, Vitamin D3 in particular, as well as defects in the Vitamin D receptor can cause rickets in humans. HVDRR can occur when the VDR is impaired in its ability to activate transcription in response to the 1,25-(OH)2D3 ligand [7]. VDR regulates the hormonal form of Vitamin D, through modifying the transcription of the target to a certain sequence of DNA, called the Vitamin D responsive element (VDRE). This activation requires an additional receptor, which is a Retinoid X Receptor (RXR) to bind to the heterodimer [7]. A mutation in the transcription of the protein has the potential to result in this disease and the mutation results in the <scene name='48/483885/Heterodimer/1'>heterodimer</scene> not forming properly. Current research shows that there are two mechanisms that can cause this failed transcription and inability to bind to the heterodimer to occur within the body. The first is a point mutation of an amino acid in the zinc finger region of the VDR that reduces the binding of the heterodimer, this region is found in the amino acid residues 21-85 [7]. The other is a premature stop codon in the DNA sequences that does not allow for the full transcription, which can have an effect of reducing the affinity of the heterodimer binding [7]. Although there may be other mutations that could cause HVDRR to occur within the body, these two types of mutations have been shown to be major causes of the disease. |
| - | <scene name='48/483885/Denv-3_mtase/2'>DENV-3 MTase</scene>
| + | |
| - | structure shows semblance to other members of the Flavivirus family as can be seen when comparing it to <scene name='48/483885/Wnv_mtase/3'>WNV MTase (West Nile Virus Methyltransferase)</scene>, and <scene name='48/483885/Denv-2_mtase/2'>DENV-2 MTase</scene>. Structural similarity especially appears within the binding pocket of these MTases regardless of SAH/SAM ligands and therefore the methylated-state of the SAM molecule shows unimportant affect over ligand-protein reciprocity. | + | |
| | | | |
| - | A unique hydrophobic pocket (viewable in <scene name='48/483885/Hydrophobic_pocket_denv-3mtase/3' caption='3p8z, hydrophobic pocket above adenine base of SAH inhbitor'>DENV-3 MTase Hydrophobic Surface Model</scene> and <scene name='48/483885/Hydrophobic_pocket_denv-2mtase/2'>DENV-2 MTase Hydrophobic Surface Model</scene>) was identified in this protein above the adenine base (of SAH) and was first identified in West Nile Virus Methyltransferase. It can be found within conserved amino acids Phe-133, Ile-147, Gly-148, Glu-149, Arg-160, Arg 163, Val-164, and Leu-182. Phe-133 and Ile-147 are of particular interest because of their functionality in the binding cavity and their formation of the hydrophobic pocket. Alanine mutations revealed various effects on the production and viral titer of DENV-3 MTase. The Table below<ref>J Biol Chem. 2011 Feb 25;286(8):6233-40. Epub 2010 Dec 8. PMID:21147775 doi:10.1074/jbc.M110.179184</ref>, shows the various effects of Alanine mutations at selected sites. These transfectation experiments revealed that mutations at the N-7 (R160A, R160A) or 2’-O (F133A, L18A) methylation activities were significantly depreciated. Melting temperatures in the table indicate low mis-folding in the mutant MTases with respect to the wild type. These attributes confirm observations in WNV MTase; that the identified hydrophobic pocket is critical to Flavivirus cap methylations and replication within cell environments. Diminishment in these properties decrease the virus’s overall potency. Therefore selective inhibition of DENV-3 MTase at this unique binding cavity becomes a viable antiviral strategy.<ref>J Biol Chem. 2011 Feb 25;286(8):6233-40. Epub 2010 Dec 8. PMID:21147775 doi:10.1074/jbc.M110.179184</ref>
| + | Also, the VDR has an effect on the hair follicle cycle, which has been observed through the eliminating the expression of this receptor. In null-VDR mice, it has been shown that with normal mineral ion levels that the mice result in alopecia, a disease that causes hair loss [8]. VDR is expressed in the hair follicle keratinocytes and its levels are higher in the late anagen and catagen stages of the hair cycle [8]. These two stages are vital in the differentiation and proliferation of hair follicle keratinocytes, which regulate hair growth in the body. Much research has been done into the mechanism in which the VDR effects the hair follicle cycle with the overall mechanism still unknown. The mechanism was first believed to be that of binding the VDR to 1,25- dihydroxyvitamin D causing transactivation due to the fact that targeted expressions of wild-type VDR to the keratinocytes of VDR null mice rescued alopecia [8]. Although, this was disproven through investigations in vitamin D3-deficient mice that had no detectable 1,25-dihydroxyvitamin D for the VDR to bind, yet the mice did not develop alopecia. This shows that the VDR transcriptional activation of DNA is not the main cause of the loss of hair follicles. Current research observes the ligand-independent actions of the VDR that have not been observed extensively as a mechanism [8]. Nuclear receptor co-repressor genes have been observed in studies to have an effect on the hair follicle cycle including the HR gene (Hairless). This corepressor has been shown to have interactions with the VDR in vivo and tests with the mutation of Hairless have caused alopecia in mice in vivo [8]. Thus, although the mechanism behind the interaction of Hairless and the VDR is still unknown it has been shown in studies that there is a relationship between the two in the body and that it has been linked to the hair follicle cycle. |
| | | | |
| - | Compounds were experimentally used to test for inhibitory strength and several key conclusions were made. SAH is non-selective and therefore inhibits eukaryotic, bacterial and viral DENV, WNV, human RNA, DNA, and histone MTases. Attachment of a benzyl ring increased selectivity for DENV MTase, however extending the aryl group by a length of one carbon reduced inhibition significantly due to hydrophobic steric hindrance, and increased substituent flexibility causing bond attenuation. Meta-substitution on the benzyl moiety caused significant increase in inhibition compared to SAH. Larger bromine and iodine groups, however, showed reduced inhibition suggesting steric hindrance from bulky groups reduces inhibition. The N6-substituted benzy ring with chloro functionality revealed rotation of Phe-133 to accommodate the bulky benzyl group, and strong cation-pi interaction between Arg-163 and the aryl-ring, the overall effect of which was tighter binding and subsequent better inhibition.<ref>J Biol Chem. 2011 Feb 25;286(8):6233-40. Epub 2010 Dec 8. PMID:21147775 doi:10.1074/jbc.M110.179184</ref> | |
| - | | |
| - | | |
| - | | |
| - | [[Image:Screen_Shot_2015-04-05_at_11.19.05_PM.png]] | |
| - | | |
| - | | |
| - | | |
| - | <br><br><br><br> | |
| | | | |
| | ==Quiz Question 1== | | ==Quiz Question 1== |
| | + | Osteoporosis is a disease in which the bones become porous and fragile. The most common cause of the ailment is calcium deficiency. As the vitamin D receptor has association with calcium uptake, mutations in VDR could be detrimental.If an individual had a point mutation that would replace <scene name='48/483885/Point_mutation/1'>His305 and His397</scene> with serine amino groups, would that individual be more likely to develop osteoporosis? why? |
| | + | ==See Also== |
| | + | *[[3w0a]] |
| | + | *[[3w0c]] |
| | + | *[[3w0y]] |
| | + | *[[3w5t]] |
| | + | *[[3w5r]] |
| | + | *[[1db1]] |
| | | | |
| - | Question1:
| + | ==Credits== |
| - | <Structure load='3p8z' size='300' frame='true' align='right' caption='3P8Z, Dengue virus Methyltransferase (MTase) bound to a SAM-based inhibitor' scene='48/483885/Initial_scene/3' />
| + | |
| - | Why are ligands <scene name='48/483885/Initial_scene/3'>S-Adenosil-homocysteine (SAH) and (S)-2-amino-4-(((2S,3S,4R,5R)-5-(6-(3-chlorobenzylamino)-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methylthio)butanoic acid, (36A)</scene> on Dengue Virus Methyltransferase predicted to have similar function of inhibiting Dengue Virus MTase?
| + | |
| | | | |
| | + | Introduction - Kate Daborowski |
| | | | |
| - | [[Image:SAH.JPG]]<br> '''SAH'''
| + | Overall Structure - Aaron Thole and Benjamin Rizkin |
| | | | |
| - | [[Image:36A.JPG]]<br> '''36A'''
| + | Drug Binding Site - Roger Crocker |
| | | | |
| | + | Additional Features - Patrick Murphy |
| | | | |
| - | A). They are both nonpolar ligands.
| + | Quiz Question 1 - Roger Crocker |
| | + | ==References== |
| | + | <references/> |
| | | | |
| - | B). They are very similar in structure. | + | 1. Rochel N, Wurtz JM, Mitschler A, Klaholz B, Moras D. The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand. Mol Cell. 2000 Jan;5(1):173-9. |
| | | | |
| - | C). They are both located on the outside of the enzyme.
| + | 2. VDR Gene http://www.genecards.org/cgi-bin/carddisp.pl?gene=VDR (accessed Apr 2, 2016). |
| | | | |
| - | D). None of the above.
| + | 3. Vitamin D3 https://pubchem.ncbi.nlm.nih.gov/compound/Vitamin_D3#section=2D-Structure (accessed Apr 10, 2016). |
| | | | |
| - | <br><br><br><br><br>
| + | 4. Strugnell, S.; Deluca, H. The Vitamin D Receptor - Structure and Transcriptional Activation. Experimental Biology and Medicine1997, 215, 223–228. |
| | | | |
| - | ==Quiz Question 2==
| + | 5. Vitamin D Receptor http://pdb101.rcsb.org/motm/155 (accessed Apr 4, 2016). |
| | | | |
| - | Question 2:
| + | 6. 3D Binding Pocket http://www.rcsb.org/pdb/explore/jmol.do?structureId=1DB1&residueNr=VDX (accessed Apr 4, 2016). |
| - | Compound 36A in question 1 showed improved inhibitory potency against DENV MTase activity with respect to SAH. Utilizing the diagram below, what would be the best explanation for such an improvement? <ref>image taken from. J Biol Chem. 2011 Feb 25;286(8):6233-40. Epub 2010 Dec 8. PMID:21147775 doi:10.1074/jbc.M110.179184</ref>
| + | |
| | | | |
| - | [[Image:Screen Shot 2015-04-05 at 11.06.41 PM.png]]
| + | 7. Whitfield, G.K.; Selznick, S.H.; Haussler, C.A.; Hsieh, J.; Galligan, M.A.; Jurutka, P.W.; Thompson, P.D.; Lee, S.M.; Zerwekh, J.E.; Haussler, M.R. Vitamin D Receptors from Patients with Resistance to 1,25-Dihydroxyvitamin D3: Point Mutations Confer Reduced Transactivation in Response to Ligand and Impair Interaction with the Retinoid X Receptor Heterodimeric Partner. Mol Endocrinol, 1996 10 (12): 1617-1631 |
| | | | |
| - | | + | 8. Skorija, K.; Cox, M.; Sisk, J.M.; Dowd, D.R.; MacDonald, P.N.; Thompson, C.C.; Demay, M.B. Ligand- Independent Actions of the Vitamin D Receptor Maintain Hair Follicle Homeostasis. Mol Endocrinol, April 2005, 19(4):855–862 |
| - | A.) Improved Hydrogen bonding
| + | |
| - | | + | |
| - | B.) Steric stability within the hydrophobic pocket
| + | |
| - | | + | |
| - | C.) Cation-pi interaction with N-6 aryl ring and R163
| + | |
| - | | + | |
| - | D.) Extension of carbon chain from the N-6 to the aryl substituent increasing flexibility and thus creating strong a stronger binding affinity for the hydrophobic pocket
| + | |
| - | | + | |
| - | E.) None of the Above
| + | |
| - | | + | |
| - | ==See Also==
| + | |
| - | *http://www.uniprot.org/uniprot/C1KBQ3
| + | |
| - | *http://www.ebi.ac.uk/pdbsum/3p8z
| + | |
| - | *http://proteopedia.org/wiki/index.php/3p8z
| + | |
| - | *http://www.proteopedia.org/wiki/index.php/3p97
| + | |
| - | *http://www.proteopedia.org/wiki/index.php/SAM-dependent_methyltransferase
| + | |
| - | | + | |
| - | ==Credits==
| + | |
| - | | + | |
| - | Introduction - Adlina Hasni, Xuanting Wang
| + | |
| - | | + | |
| - | Overall Structure - Michael Bresnahan
| + | |
| - | | + | |
| - | Drug Binding Site - Jon Dullea
| + | |
| - | | + | |
| - | Additional Features - Matthew Caissy
| + | |
| - | | + | |
| - | Quiz Question 1 - Xuanting Wang
| + | |
| - | | + | |
| - | Quiz Question 2 - Matthew Caissy
| + | |
| - | | + | |
| - | ==References==
| + | |
| - | <references/>
| + | |
by Roger Crocker, Kate Daborowski, Patrick Murphy, Benjamin Rizkin and Aaron Thole
|
Introduction
The (VDR) is a ligand-dependent transcriptional regulator with two strands. VDR belongs to the superfamily of nuclear receptors which control homeostasis, cell differentiation and growth, and many physiological processes. All proteins that belong to the nuclear receptor superfamily have a variable N-terminus region (A/B region), a hinge region that is flexible (D region), a conserved DNA-binding region (DBD, C region), and a moderately conserved ligand-binding region (LBD, E/F region). In the case of VDR, the A/B region is very short so it does not have any AF-1 function and the ligand binding region has a dimerization interface and a transcriptional activation domain that is ligand-dependent (AF-2).[1]
The VDR has both an active and suppressed form. The activation or suppression function is caused by the binding of the DR3 response element as a heterodimer with the retinoid X receptor of the target genes. Due to the interactions with the basal transcriptional machinery and transcriptional cofactors, transcription is either activated or suppressed. When VDR is in its active form it regulates both phosphate and calcium metabolism, has immunosuppressive effects, and induces cell differentiation. When there are defects in the VDR that effect its metabolism it can lead to diseases such as severe rickets, secondary hyperparathyroidism, and hypocalcemia. Though defects in VDR can cause many diseases, fully functioning VDR can be used as treatment for disease such as cancer, autoimmune disease, psoriasis, osteoporosis, and renal osteodystrophy.[1]
Overall Structure
The contains 427 amino acids with a total molecular weight of 48,289 Da. The protein is also composed almost entirely of alpha helices with only a single beta sheet. The vitamin D receptor also does not have a quaternary structure [2]. is a large organic compound made up of 27 carbon atoms, 44 hydrogen atoms and a single oxygen atom, with the ligand having a total molecular weight of 385 Da [3]. In studying the vitamin d receptor, the regions of the protein have been categorized into domains, with the A/B domain located at the N-terminus, the C domain, which is located between amino acid 20 and amino acid 115, the D domain, which is located between the end of the C domain and amino acid 220, and the EF domain, which encompasses the rest of the protein [4].
Other aspects of interest about the vitamin D receptor include the protein revealing a binding pocket when it is in its active folded state, allowing the ligand to bind to the receptor. The ligand interacts with the activation helix by stabilizing the agonist position. This is accomplished through Van der Waals interactions between the ligand and the activation helix. The activation ligand is a nuclear receptor. There is also some empty space observed around the aliphatic chain [1].
The protein has an active conformation of 1,25 (OH)_2D_3 that has a ligand binding pocket in its active folded state. The activation ligand, a nuclear receptor (VDR), interacts with with the activation helix by stabilizing the agonist position. This is accomplished through Van der Waals interactions between the ligand and the activation helix. There is some empty space observed around the aliphatic chain, indicating the presence of water to stabilize all possible hydrogen bonds [1].
Binding Interactions
Protein 1db1 is found to complex with 1,25 Dihydroxy . This molecule has three notable alcohol groups shown in red. Oxygen is electronegative, giving alcohols the ability to participate in hydrogen bonding with the protein. Vitamin D3 has a large number of relations with the residues on the protein chain. First in the sequence are . Tyr143, shown in blue, is the closest to the ligand at 2.83 angstroms. This is sightly large but there is still the possibility of hydrogen bonding. Tyr147 in green and Phe150 in black are also known to have interactions with vitamin D3 they are farther away and therefore less significant. Next down the peptide chain are . Ser237, shown in green, has significant interactions with vitamin D3 this can be seen by its short distance 2.78 angstroms. Only 40 residues away, more hydrogen bonding is occurring. form bonds with the same oxygen atoms as Tyr143 and Ser237 respectively. This means that the oxygen atoms of vitamin D3, when bound to the receptor, are negatively charged and stabilized by protons. On the opposite end of vitamin D3 there is an additional negatively charged oxygen. Although this oxygen does not participate in hydrogen bonding. , shown in blue and green respectively, Contain aromatic rings. These rings are able to momentarily accept the electrons donated by the oxygen because they can delocalize the charge. This creates two pseudo-covalent bonds that is approximately 2.81 angstroms. When looking at , it can be seen that all of the binding sights are centered around oxygen [5,6].
Additional Features
Hereditary Vitamin D Resistant Rickets (HVDRR) is a condition that most commonly occurs in children and results in soft and weak bone formation, which often causes deformities in bone structure. A lack of proper nutrients, Vitamin D3 in particular, as well as defects in the Vitamin D receptor can cause rickets in humans. HVDRR can occur when the VDR is impaired in its ability to activate transcription in response to the 1,25-(OH)2D3 ligand [7]. VDR regulates the hormonal form of Vitamin D, through modifying the transcription of the target to a certain sequence of DNA, called the Vitamin D responsive element (VDRE). This activation requires an additional receptor, which is a Retinoid X Receptor (RXR) to bind to the heterodimer [7]. A mutation in the transcription of the protein has the potential to result in this disease and the mutation results in the not forming properly. Current research shows that there are two mechanisms that can cause this failed transcription and inability to bind to the heterodimer to occur within the body. The first is a point mutation of an amino acid in the zinc finger region of the VDR that reduces the binding of the heterodimer, this region is found in the amino acid residues 21-85 [7]. The other is a premature stop codon in the DNA sequences that does not allow for the full transcription, which can have an effect of reducing the affinity of the heterodimer binding [7]. Although there may be other mutations that could cause HVDRR to occur within the body, these two types of mutations have been shown to be major causes of the disease.
Also, the VDR has an effect on the hair follicle cycle, which has been observed through the eliminating the expression of this receptor. In null-VDR mice, it has been shown that with normal mineral ion levels that the mice result in alopecia, a disease that causes hair loss [8]. VDR is expressed in the hair follicle keratinocytes and its levels are higher in the late anagen and catagen stages of the hair cycle [8]. These two stages are vital in the differentiation and proliferation of hair follicle keratinocytes, which regulate hair growth in the body. Much research has been done into the mechanism in which the VDR effects the hair follicle cycle with the overall mechanism still unknown. The mechanism was first believed to be that of binding the VDR to 1,25- dihydroxyvitamin D causing transactivation due to the fact that targeted expressions of wild-type VDR to the keratinocytes of VDR null mice rescued alopecia [8]. Although, this was disproven through investigations in vitamin D3-deficient mice that had no detectable 1,25-dihydroxyvitamin D for the VDR to bind, yet the mice did not develop alopecia. This shows that the VDR transcriptional activation of DNA is not the main cause of the loss of hair follicles. Current research observes the ligand-independent actions of the VDR that have not been observed extensively as a mechanism [8]. Nuclear receptor co-repressor genes have been observed in studies to have an effect on the hair follicle cycle including the HR gene (Hairless). This corepressor has been shown to have interactions with the VDR in vivo and tests with the mutation of Hairless have caused alopecia in mice in vivo [8]. Thus, although the mechanism behind the interaction of Hairless and the VDR is still unknown it has been shown in studies that there is a relationship between the two in the body and that it has been linked to the hair follicle cycle.
Quiz Question 1
Osteoporosis is a disease in which the bones become porous and fragile. The most common cause of the ailment is calcium deficiency. As the vitamin D receptor has association with calcium uptake, mutations in VDR could be detrimental.If an individual had a point mutation that would replace with serine amino groups, would that individual be more likely to develop osteoporosis? why?
See Also
Credits
Introduction - Kate Daborowski
Overall Structure - Aaron Thole and Benjamin Rizkin
Drug Binding Site - Roger Crocker
Additional Features - Patrick Murphy
Quiz Question 1 - Roger Crocker
References
- ↑ Rochel N, Wurtz JM, Mitschler A, Klaholz B, Moras D. The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand. Mol Cell. 2000 Jan;5(1):173-9. PMID:10678179
1. Rochel N, Wurtz JM, Mitschler A, Klaholz B, Moras D. The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand. Mol Cell. 2000 Jan;5(1):173-9.
2. VDR Gene http://www.genecards.org/cgi-bin/carddisp.pl?gene=VDR (accessed Apr 2, 2016).
3. Vitamin D3 https://pubchem.ncbi.nlm.nih.gov/compound/Vitamin_D3#section=2D-Structure (accessed Apr 10, 2016).
4. Strugnell, S.; Deluca, H. The Vitamin D Receptor - Structure and Transcriptional Activation. Experimental Biology and Medicine1997, 215, 223–228.
5. Vitamin D Receptor http://pdb101.rcsb.org/motm/155 (accessed Apr 4, 2016).
6. 3D Binding Pocket http://www.rcsb.org/pdb/explore/jmol.do?structureId=1DB1&residueNr=VDX (accessed Apr 4, 2016).
7. Whitfield, G.K.; Selznick, S.H.; Haussler, C.A.; Hsieh, J.; Galligan, M.A.; Jurutka, P.W.; Thompson, P.D.; Lee, S.M.; Zerwekh, J.E.; Haussler, M.R. Vitamin D Receptors from Patients with Resistance to 1,25-Dihydroxyvitamin D3: Point Mutations Confer Reduced Transactivation in Response to Ligand and Impair Interaction with the Retinoid X Receptor Heterodimeric Partner. Mol Endocrinol, 1996 10 (12): 1617-1631
8. Skorija, K.; Cox, M.; Sisk, J.M.; Dowd, D.R.; MacDonald, P.N.; Thompson, C.C.; Demay, M.B. Ligand- Independent Actions of the Vitamin D Receptor Maintain Hair Follicle Homeostasis. Mol Endocrinol, April 2005, 19(4):855–862
|