User:Braden Sciarra/Sandbox 1
From Proteopedia
< User:Braden Sciarra(Difference between revisions)
| (5 intermediate revisions not shown.) | |||
| Line 2: | Line 2: | ||
<StructureSection load='1F8I' size='340' side='right' caption='Isocitrate Lyase from ''Mycobacterium tuberculosis''' scene=''> | <StructureSection load='1F8I' size='340' side='right' caption='Isocitrate Lyase from ''Mycobacterium tuberculosis''' scene=''> | ||
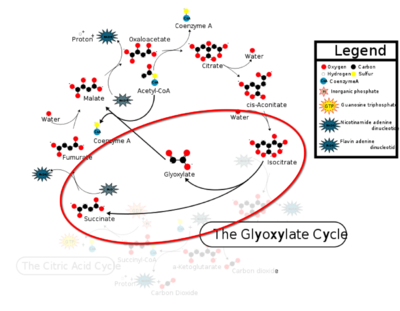
[[Image:CAC.png|400 px|right|thumb|Figure 1: ICL mediated glyoxylate shunt pathway of the Citric Acid Cycle]] | [[Image:CAC.png|400 px|right|thumb|Figure 1: ICL mediated glyoxylate shunt pathway of the Citric Acid Cycle]] | ||
| - | [http://en.wikipedia.org/wiki/Isocitrate_lyase Isocitrate Lyase] (ICL) is a metabolic enzyme that converts the metabolite isocitrate into glyoxylate and succinate. ICL is a homotetramer with each monomer being composed of 14 alpha helices, 14 beta sheets, and a magnesium ion cofactor. ICL has shown clinical relevance in the disease state [http://en.wikipedia.org/wiki/Tuberculosis Tuberculosis] where it is responsible for the persistence of Mycobacterium tuberculosis during the chronic stage of infection | + | [http://en.wikipedia.org/wiki/Isocitrate_lyase Isocitrate Lyase] (ICL) is a metabolic enzyme that converts the metabolite isocitrate into glyoxylate and succinate. ICL is a homotetramer with each monomer being composed of 14 alpha helices, 14 beta sheets, and a magnesium ion cofactor. ICL has shown clinical relevance in the disease state [http://en.wikipedia.org/wiki/Tuberculosis Tuberculosis] where it is responsible for the persistence of Mycobacterium tuberculosis during the chronic stage of infection<ref name="genes">PMID: 18054522</ref> This survival strategy mediated by ICL is characterized by a metabolic shortcut within the [http://en.wikipedia.org/wiki/Citric_acid_cycle Citric Acid Cycle]. ICL creates this shunt pathway by converting isocitrate to succinate and glyoxylate, diverting acetyl-CoA from the beta-oxidation of fatty acids<ref name="ICL">PMID:10932251</ref><ref name="ICL2">PMID: 2696959</ref>. |
== Structure == | == Structure == | ||
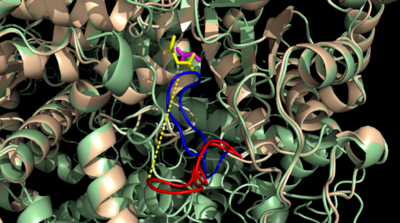
[[Image:homotetramer.png|150 px|left|thumb|Figure 2: C2 Symmetry of the homotetramer isocitrate lyase]] | [[Image:homotetramer.png|150 px|left|thumb|Figure 2: C2 Symmetry of the homotetramer isocitrate lyase]] | ||
| - | The ICL homotetramer possesses C2 symmetry, with an axis of rotation at x-axis, y-axis, and z-axis of the enzyme. Two individual subunits off ICL are held together by a characteristic <scene name='69/697526/Helix_swapping/3'>Helix Swapping</scene> between three alpha helices formed by residues 370-384, 349-367, and 399-409 on neighboring monomers<ref name="ICL">PMID:10932251</ref>. The interlocking mechanism created by these helices provides additional strength to hold the two monomeric subunits together, allowing ICL to essentially be composed of two dimerized subunits<ref name="ICL2" | + | The ICL homotetramer possesses C2 symmetry, with an axis of rotation at x-axis, y-axis, and z-axis of the enzyme. Two individual subunits off ICL are held together by a characteristic <scene name='69/697526/Helix_swapping/3'>Helix Swapping</scene> between three alpha helices formed by residues 370-384, 349-367, and 399-409 on neighboring monomers<ref name="ICL">PMID:10932251</ref>. The interlocking mechanism created by these helices provides additional strength to hold the two monomeric subunits together, allowing ICL to essentially be composed of two dimerized subunits<ref name="ICL2"/>. This interaction will bury approximately 18% of the surface of each subunit, and will help to shield the interior binding site from hydration. |
== Active Site == | == Active Site == | ||
| Line 14: | Line 14: | ||
===Catalytic Loop=== | ===Catalytic Loop=== | ||
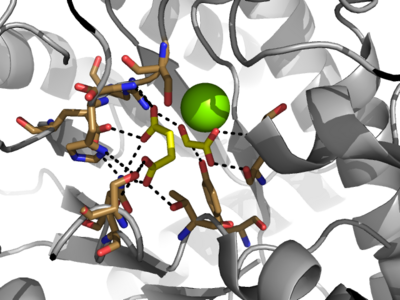
[[Image:Openvsclosed..png|400 px|left|thumb|Figure 3: Highly ordered hydrogen bonding network within the active site of ICL]] | [[Image:Openvsclosed..png|400 px|left|thumb|Figure 3: Highly ordered hydrogen bonding network within the active site of ICL]] | ||
| - | The <scene name='69/697526/Catalytic_loop/1'>Catalytic Loop</scene> of the Isocitrate Lyase enzyme is composed of residues 185-196, and can exist in both the open and closed conformation. In the open conformation, the catalytic loop is oriented such that the catalytic CYS191 residue is located far from the active site, allowing for solvent accessibility and substrate binding. Upon substrate binding, the catalytic loop adopts a closed conformation, moving between ten and fifteen angstroms<ref name="ICL">PMID:10932251</ref>. This closed conformation will cause the binding site to become inaccessible to the solvent. The loop closure is triggered by the movement of the Mg2+ ion that occurs upon binding of the succinate. This movement of the Mg2+ ion results in electrostatic interactions at LYS189, causing the loop to close. | + | The <scene name='69/697526/Catalytic_loop/1'>Catalytic Loop</scene> of the Isocitrate Lyase enzyme is composed of residues 185-196, and can exist in both the open and closed conformation. In the open conformation, the catalytic loop is oriented such that the catalytic CYS191 residue is located far from the active site, allowing for solvent accessibility and substrate binding.<ref name="solvent">Connely, M. L. Solvent-accessible surfaces of proteins and nucleic acids "Science" 221:709-713 (1983). DOI: 10.1126/science.6879170</ref> Upon substrate binding, the catalytic loop adopts a closed conformation, moving between ten and fifteen angstroms<ref name="ICL">PMID:10932251</ref>. This closed conformation will cause the binding site to become inaccessible to the solvent. The loop closure is triggered by the movement of the Mg2+ ion that occurs upon binding of the succinate. This movement of the Mg2+ ion results in electrostatic interactions at LYS189, causing the loop to close. |
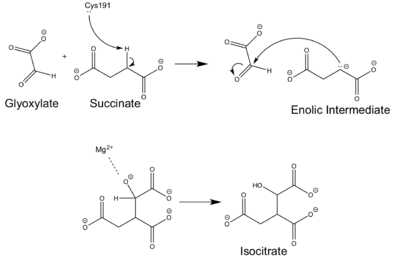
==Mechanism== | ==Mechanism== | ||
Current revision
Isocitrate Lyase from Mycobacterium Tuberculosis
| |||||||||||
References
- ↑ Srivastava V, Jain A, Srivastava BS, Srivastava R. Selection of genes of Mycobacterium tuberculosis upregulated during residence in lungs of infected mice. Tuberculosis (Edinb). 2008 May;88(3):171-7. Epub 2007 Dec 3. PMID:18054522 doi:http://dx.doi.org/10.1016/j.tube.2007.10.002
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 Sharma V, Sharma S, Hoener zu Bentrup K, McKinney JD, Russell DG, Jacobs WR Jr, Sacchettini JC. Structure of isocitrate lyase, a persistence factor of Mycobacterium tuberculosis. Nat Struct Biol. 2000 Aug;7(8):663-8. PMID:10932251 doi:10.1038/77964
- ↑ 3.0 3.1 3.2 Beeching JR. High sequence conservation between isocitrate lyase from Escherichia coli and Ricinus communis. Protein Seq Data Anal. 1989 Dec;2(6):463-6. PMID:2696959
- ↑ 4.0 4.1 4.2 4.3 Masamune et al. Bio-Claisen condensation catalyzed by thiolase from Zoogloea ramigera. Active site cysteine residues. "Journal of the American Chemical Society" 111: 1879-1881 (1989). DOI: 10.1021/ja00187a053
- ↑ Connely, M. L. Solvent-accessible surfaces of proteins and nucleic acids "Science" 221:709-713 (1983). DOI: 10.1126/science.6879170