This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 1067
From Proteopedia
(Difference between revisions)
| (21 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | ==Zinc Transporter Yiip== | |
| - | == | + | <StructureSection load='3h90' size='350' side='right' caption='Escherichia coli reca protein-bound DNA (PDB entry [[3h90]])' scene=''> |
| - | <StructureSection load=' | + | |
| - | + | ||
| - | + | ||
| - | = | + | <scene name='69/694234/3h90/2'>3h90</scene> is an integral membrane protein of the E. coli bacteria that regulates the import and export of Zn<sup>2+</sup>. This regulation prevents zinc toxicity and deprivation within the cytoplasm, which can lead to cell death. |
| - | + | ||
| - | + | ||
| - | == | + | == Structure == |
| + | Yiip's complete protein structure is identified as a dimer. Each monomer structure is classified into two different domains, the Trans-Membrane Domain (TMD) and C-Terminus Domain (CTD). The TMD consists of six helices, forming binding site A, four of which are oriented in a parallel manner, with respect to each other, while the remaining two are aligned anti parallel to this four helix cluster. A "<scene name='69/694234/Bridge/1'>salt bridge</scene>" or "charge interlock" consisting of four amino acid residues, two Lysines (Lys 77) and two Aspartates (Asp 207), forms a junction that both monomers of Yiip converge at, forming a pivot point for conformation changes. A large portion of the protein containing binding site C, approximately 30 Å in length<sup>[1]</sup>, protrudes into the cytoplasm functioning as a zinc sensor within the cell. | ||
| - | == | + | == Salt Bridge == |
| + | |||
| + | == Electrostatic Interactions == | ||
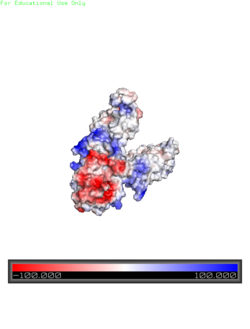
| + | [[Image:Yiip_Electrostatic.png|250px|left|thumb|Electrostatic Charge Distribution]]<scene name='69/694234/Electro/1'>Charge distribution</scene> along the exterior surface of the protein is primarily neutral for the TMDs, but transitions to positive near the location of the charge interlock and interior side of the cell membrane. This positive section is characteristic of trans-membrane proteins as a means of achieving proper orientation within the cell membrane. Binding sites A, B, and C, as well as the CTDs of both monomers, all possess a high negative charge relative to the other charges present, facilitating the binding and releasing of Zn<sup>2+</sup> ions. The two CTDs are held together by the charge interlock and hydrophobic interactions of the TMDs despite their electrostatic repulsion. Upon the release of Zn<sup>2+</sup> ions, the CTDs undergo electronegativity alterations, which enables domain separation. | ||
| + | |||
| + | == Conformation Changes == | ||
| - | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
</StructureSection> | </StructureSection> | ||
| + | |||
== References == | == References == | ||
<references/> | <references/> | ||
Current revision
Zinc Transporter Yiip
| |||||||||||