We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
JMS/Sandbox25
From Proteopedia
(Difference between revisions)
| (15 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
==Your Heading Here (maybe something like 'Structure')== | ==Your Heading Here (maybe something like 'Structure')== | ||
<StructureSection load='Conevo_1.pdb' size='340' side='right' caption='Caption for this structure' scene='70/700369/Golden_comp_1/1'> | <StructureSection load='Conevo_1.pdb' size='340' side='right' caption='Caption for this structure' scene='70/700369/Golden_comp_1/1'> | ||
| - | This is a default text for your page '''JMS/Sandbox25'''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
| - | == | + | == Structural highlights == |
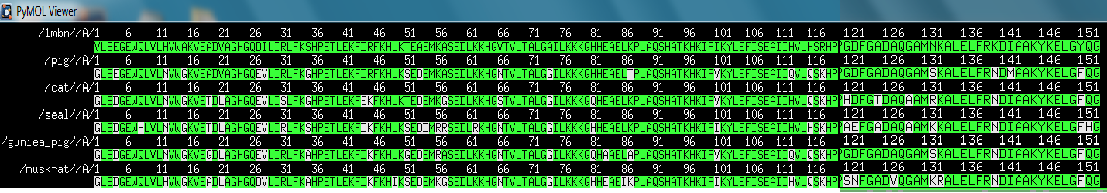
| - | == | + | key: w = whale; p = pig; s = seal; c = cat; m = muskrate; g = guniae pig. |
| - | = | + | <scene name='70/700369/Golden_comp_1/2'>all areas with possible change in net charge</scene> 8,12,31,53,57,82,84,87,113,116,121,122,132,140. |
| - | + | <scene name='70/700369/Golden_comp_1/3'>only times created a hydrophobic patch</scene> was w53p, c121s and g122m. in every other case removing a hydrophobic patch in each case! | |
| - | <scene name='70/700369/Golden_comp_1/ | + | <scene name='70/700369/Golden_comp_1/4'>funny at 84</scene>, muskrat lost a hydrophic patch but becomes less charged. |
| - | + | <scene name='70/700369/Golden_comp_1/6'>could have increased the charge AND the net charge at once ... but didn't!</scene> in blue. in red, is a conserved negatively charge aa - could been both or just increase in charge. | |
| + | <scene name='70/700369/Golden_comp_1/7'>showing lost hydrophobic patch, increase in charge of muskrat 84 in purple, and the three increase in hydophobic patches in green</scene> | ||
| + | |||
| + | p:w 12,53,87,113,116 | ||
| + | c:s 8,31,57,82,116,121 | ||
| + | g:m 12,82,84,113,122,132 | ||
| + | |||
| + | wm 12,113 | ||
| + | sm 82 | ||
| + | |||
| + | <scene name='70/700369/Golden_comp_1/9'>showing where each evolution step occurred by aquatic mammal.</scene> | ||
| + | |||
| + | <scene name='70/700369/Globin/1'>dude!!</scene>a backbone of 3 myoglobin pairs, oxy and deoxy wt hemoglobin, and deoxy sickle cell, and the horse myoglobin dimer. | ||
| + | |||
| + | showing 1mbn with the sites that have evolved. and hbB from sickle, with the e6v in halo. <scene name='70/700369/Globin/2'>funny no i6+ in aquatic mammals.</scene> | ||
| + | |||
| + | w whale -> r c cat -> q s seal -> N g gunea pig -> o p pig m muskrat | ||
| + | z oaa oxy y oab | ||
| + | x doa deoxy w dob v doc u dod | ||
| + | a-h ds{a-h} sickle cell | ||
| + | t horse a s horse b | ||
| + | |||
| + | <scene name='70/700369/Globin/5'>whale horse comparison</scene> | ||
| + | |||
| + | <scene name='70/700369/Globin/6'>whale to horse comp. emphasizing halves of protein</scene>. next up is interface. | ||
| + | |||
| + | note the v to i mutation in all three pairs! why?? also, horse is i.. | ||
</StructureSection> | </StructureSection> | ||
| + | |||
| + | pymol sequence alignment | ||
| + | [[Image:Comp_seq_1.png]] | ||
== References == | == References == | ||
<references/> | <references/> | ||
Current revision
Your Heading Here (maybe something like 'Structure')
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644