Journal:Cell:1
From Proteopedia
(Difference between revisions)

| (3 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='' size='400' side='right' scene='User:David_Canner/Workbench/Opening/2' caption='Solved Structures of IFNAR1/2/IFN Complexes, [[1dq8]]'> | |
=== Structural linkage between ligand discrimination and receptor activation by type I interferons === | === Structural linkage between ligand discrimination and receptor activation by type I interferons === | ||
<big>Christoph Thomas, Ignacio Moraga, Doron Levin, Peter O. Krutzik, Yulia Podoplelova, Angelica Trejo, Choongho Lee, Ganit Yarden, Susan E. Vleck, Jeffrey S. Glenn, Garry P. Nolan, Jacob Piehler, Gideon Schreiber, K. Christopher Garcia</big><ref>DOI 10.1016/j.cell.2011.06.048</ref> | <big>Christoph Thomas, Ignacio Moraga, Doron Levin, Peter O. Krutzik, Yulia Podoplelova, Angelica Trejo, Choongho Lee, Ganit Yarden, Susan E. Vleck, Jeffrey S. Glenn, Garry P. Nolan, Jacob Piehler, Gideon Schreiber, K. Christopher Garcia</big><ref>DOI 10.1016/j.cell.2011.06.048</ref> | ||
| Line 12: | Line 12: | ||
====IFNAR2-IFN interaction==== | ====IFNAR2-IFN interaction==== | ||
[[Image:IFNa_IFNAR2_interaction_map.png|300px||right|]] | [[Image:IFNa_IFNAR2_interaction_map.png|300px||right|]] | ||
| + | {{Clear}} | ||
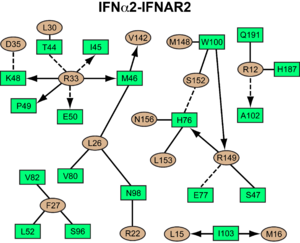
<scene name='User:David_Canner/Workbench/Opening_ifna/2'>Interferon</scene> interacts primarily with the <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_interaction/1'>D1 domain of IFNAR2</scene>. Arg33(IFN) appears to be the <scene name='User:David_Canner/Workbench2/Ifn_arg_33/1'>single most important residue</scene> for the interaction of the IFN ligand with IFNAR2. It forms an extensive hydrogen-bonding network with the main chain carbonyl oxygen atoms of Ile45(IFNAR2) and Glu50(IFNAR2) and the side chain of Thr44(IFNAR2). This residue is present in IFNa, IFNw, IFNb and IFNe. Two hydrophobic interaction clusters are part of the IFNa-IFNAR2 interface: the first one is formed between Leu15 and Met16 of the IFN molecule and Trp100 and Ile103 of IFNAR2; the second one comprises Leu26, Phe27, Leu30 and Val142 of the ligand and Met46, Leu52, Val80 and the methyl group of Thr44 of the receptor. Replacing <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_leu_30/1'>Leu30(IFN) with alanine</scene> reduces affinity by three orders of magnitude (the second most important residue for binding). This is surprising, as it is not engaged in any intimate contacts with IFNAR2 residues. One reason for its importance might be a <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_arg_stabilized/1'>stabilizing effect on the position of Arg33(IFN)</scene>. | <scene name='User:David_Canner/Workbench/Opening_ifna/2'>Interferon</scene> interacts primarily with the <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_interaction/1'>D1 domain of IFNAR2</scene>. Arg33(IFN) appears to be the <scene name='User:David_Canner/Workbench2/Ifn_arg_33/1'>single most important residue</scene> for the interaction of the IFN ligand with IFNAR2. It forms an extensive hydrogen-bonding network with the main chain carbonyl oxygen atoms of Ile45(IFNAR2) and Glu50(IFNAR2) and the side chain of Thr44(IFNAR2). This residue is present in IFNa, IFNw, IFNb and IFNe. Two hydrophobic interaction clusters are part of the IFNa-IFNAR2 interface: the first one is formed between Leu15 and Met16 of the IFN molecule and Trp100 and Ile103 of IFNAR2; the second one comprises Leu26, Phe27, Leu30 and Val142 of the ligand and Met46, Leu52, Val80 and the methyl group of Thr44 of the receptor. Replacing <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_leu_30/1'>Leu30(IFN) with alanine</scene> reduces affinity by three orders of magnitude (the second most important residue for binding). This is surprising, as it is not engaged in any intimate contacts with IFNAR2 residues. One reason for its importance might be a <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_arg_stabilized/1'>stabilizing effect on the position of Arg33(IFN)</scene>. | ||
Most of the residues involved in the IFNa2-IFNAR2 interaction are also found in the IFNw-IFNAR2 interface of the IFNw ternary complex. | Most of the residues involved in the IFNa2-IFNAR2 interaction are also found in the IFNw-IFNAR2 interface of the IFNw ternary complex. | ||
| - | [[Image:IFNw_IFNAR2_interaction_map.png|300px|left|]] | + | [[Image:IFNw_IFNAR2_interaction_map.png|300px|left|]] |
| + | {{Clear}} | ||
A significant difference in the IFNAR2 interface between <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_interaction_dif/5'>IFNa2</scene> and IFNw is related to <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_interaction_salt/1'>Arg149 in IFNa2</scene>, which is replaced with Lys152 in <scene name='User:David_Canner/Workbench2/Ifnw_ifnar21_structure/3'>IFNw</scene>. In the <scene name='User:David_Canner/Workbench2/Ifnw_ifnar2_interface/3'>IFNw-IFNAR2 interface</scene>, this residue forms an <scene name='User:David_Canner/Workbench2/Ifnw_ifnar2_salt/1'>intramolecular salt bridge</scene> with Glu149(IFN), but <scene name='User:David_Canner/Workbench2/Ifnw_ifnar2_no_interact/1'>does not contact Glu77 of the receptor</scene>. | A significant difference in the IFNAR2 interface between <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_interaction_dif/5'>IFNa2</scene> and IFNw is related to <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_interaction_salt/1'>Arg149 in IFNa2</scene>, which is replaced with Lys152 in <scene name='User:David_Canner/Workbench2/Ifnw_ifnar21_structure/3'>IFNw</scene>. In the <scene name='User:David_Canner/Workbench2/Ifnw_ifnar2_interface/3'>IFNw-IFNAR2 interface</scene>, this residue forms an <scene name='User:David_Canner/Workbench2/Ifnw_ifnar2_salt/1'>intramolecular salt bridge</scene> with Glu149(IFN), but <scene name='User:David_Canner/Workbench2/Ifnw_ifnar2_no_interact/1'>does not contact Glu77 of the receptor</scene>. | ||
====IFNAR1-IFN interaction==== | ====IFNAR1-IFN interaction==== | ||
| - | Because of the lower resolution of the IFNa ternary complex, we focused on the <scene name='User:David_Canner/Workbench2/Ifnw_ifnar21_structure/3'>IFNw complex</scene> in our analysis of the IFN-IFNAR1 interface. In the <scene name='User:David_Canner/Workbench2/Ifnw_ifnar21_structure/3'>IFNw-IFNAR1 complex</scene>, the <scene name='User:David_Canner/Workbench2/Ifnw_ifnar21_zoomed/1'>ligand-binding site of IFNAR1</scene> only contains two hotspot residues we could experimentally confirm, <scene name='User:David_Canner/Workbench2/W-1-tyr_70/1'>Tyr70(IFNAR1)</scene> and Phe238(IFNAR1). Substituting these residues by alanine reduces the affinity to all tested IFN ligands by more than 10-fold. On IFNw, mutation studies have shown that a charge-reversal mutation of Arg123 (Arg 120 on IFNa) leads to a total loss of activity. [[Image:IFNw_IFNAR1_interaction_map.png|300px||left|]] Indeed, this residue forms a salt bridge with Asp132(IFNAR1) in addition to a hydrogen bond with Ser182(IFNAR1). Substitution of glutamate for Arg123(IFN) would lead to electrostatic repulsion with Asp132(IFNAR1). | + | Because of the lower resolution of the IFNa ternary complex, we focused on the <scene name='User:David_Canner/Workbench2/Ifnw_ifnar21_structure/3'>IFNw complex</scene> in our analysis of the IFN-IFNAR1 interface. In the <scene name='User:David_Canner/Workbench2/Ifnw_ifnar21_structure/3'>IFNw-IFNAR1 complex</scene>, the <scene name='User:David_Canner/Workbench2/Ifnw_ifnar21_zoomed/1'>ligand-binding site of IFNAR1</scene> only contains two hotspot residues we could experimentally confirm, <scene name='User:David_Canner/Workbench2/W-1-tyr_70/1'>Tyr70(IFNAR1)</scene> and Phe238(IFNAR1). Substituting these residues by alanine reduces the affinity to all tested IFN ligands by more than 10-fold. On IFNw, mutation studies have shown that a charge-reversal mutation of Arg123 (Arg 120 on IFNa) leads to a total loss of activity. [[Image:IFNw_IFNAR1_interaction_map.png|300px||left|]] |
| + | {{Clear}} | ||
| + | Indeed, this residue forms a salt bridge with Asp132(IFNAR1) in addition to a hydrogen bond with Ser182(IFNAR1). Substitution of glutamate for Arg123(IFN) would lead to electrostatic repulsion with Asp132(IFNAR1). | ||
The low affinity of IFNAR1 for the ligand appears to be functionally relevant, as weak binding to IFNAR1 is conserved between all alpha IFNs. Three amino acid substitutions on IFNa2 at positions His57, Glu58 and Ser61 to alanine or to Tyr, Asn, and Ser, respectively, confer tighter binding to IFNAR1, but leave the affinity to IFNAR2 essentially unaltered. | The low affinity of IFNAR1 for the ligand appears to be functionally relevant, as weak binding to IFNAR1 is conserved between all alpha IFNs. Three amino acid substitutions on IFNa2 at positions His57, Glu58 and Ser61 to alanine or to Tyr, Asn, and Ser, respectively, confer tighter binding to IFNAR1, but leave the affinity to IFNAR2 essentially unaltered. | ||
| Line 32: | Line 36: | ||
The low-affinity receptor chain, IFNAR1, also <scene name='User:David_Canner/Workbench3/Morph_4/4'>undergoes major conformational changes</scene> upon ligand binding. When using D1 as anchor, D3 is moving inwards (toward the ligand) by ~15 Å. This would generate an even larger movement of the membrane-proximal SD4 domain and the transmembrane helix. The conformational changes in IFNAR1 are necessary to form the full spectrum of interactions with the IFN ligand, and to form a stable signaling complex that is able to instigate downstream signaling. In contrast to SD3, SD4 seems to be highly flexible (even more than D2 of IFNAR2). One might suggest that the conformational changes in IFNAR1 by itself will be responsible for a reduced binding affinity of IFNAR1 and may slow down the rate of ligand association to IFNAR1 directly from solution. | The low-affinity receptor chain, IFNAR1, also <scene name='User:David_Canner/Workbench3/Morph_4/4'>undergoes major conformational changes</scene> upon ligand binding. When using D1 as anchor, D3 is moving inwards (toward the ligand) by ~15 Å. This would generate an even larger movement of the membrane-proximal SD4 domain and the transmembrane helix. The conformational changes in IFNAR1 are necessary to form the full spectrum of interactions with the IFN ligand, and to form a stable signaling complex that is able to instigate downstream signaling. In contrast to SD3, SD4 seems to be highly flexible (even more than D2 of IFNAR2). One might suggest that the conformational changes in IFNAR1 by itself will be responsible for a reduced binding affinity of IFNAR1 and may slow down the rate of ligand association to IFNAR1 directly from solution. | ||
| + | |||
| + | Coordinates and structure factors have been deposited within the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB) under accession codes [[3s98]] (IFNAR1ΔSD4), [[3s8w]] (IFNAR2-D2), [[3s9d]] (binary IFNα-IFNAR2 complex), [[3se4]] (ternary IFNω complex), and [[3se3]] (ternary IFNα complex). | ||
__NOTOC__ | __NOTOC__ | ||
</StructureSection> | </StructureSection> | ||
Current revision
| |||||||||||
- ↑ Thomas C, Moraga I, Levin D, Krutzik PO, Podoplelova Y, Trejo A, Lee C, Yarden G, Vleck SE, Glenn JS, Nolan GP, Piehler J, Schreiber G, Garcia KC. Structural Linkage between Ligand Discrimination and Receptor Activation by Type I Interferons. Cell. 2011 Aug 19;146(4):621-32. PMID:21854986 doi:10.1016/j.cell.2011.06.048
Proteopedia Page Contributors and Editors (what is this?)
Christoph Thomas, Jaime Prilusky, Joel L. Sussman, Michal Harel, Alexander Berchansky
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.