User:Rana Saad/The human GABAb receptor
From Proteopedia
< User:Rana Saad(Difference between revisions)
| (140 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| + | <StructureSection load='4mqe' size='340' side='right' caption='GABAb receptor' scene=''> | ||
[[Image:Gabab.xu2014.png |thumb|400px]] | [[Image:Gabab.xu2014.png |thumb|400px]] | ||
| - | [[Image:GABAb.receptor.cartoon2.png| thumb|300px]] | ||
=='''''Introduction'''''== | =='''''Introduction'''''== | ||
| - | + | [https://en.wikipedia.org/wiki/Gamma-Aminobutyric_acid γ-Aminobutyric acid (GABA)] is the major inhibitory [http://en.wikipedia.org/wiki/Neurotransmitter neurotransmitter] in the central nervous system (CNS). It plays a key role in modulating neuronal activity by binding to specific transmembrane receptors ([http://en.wikipedia.org/wiki/GABAA_receptor GABA<sub>A</sub>],GABA<sub>B</sub> and [http://en.wikipedia.org/wiki/GABAA-rho_receptor GABA<sub>C</sub>]) in the plasma membrane of both pre- and postsynaptic neuronal level. | |
| - | + | ||
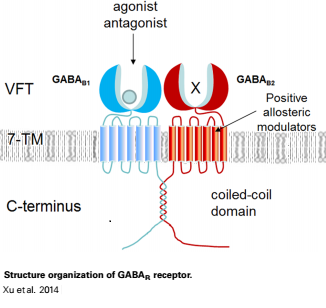
| - | + | The mammalian '''GABA<sub>B</sub>''' receptor is a class C [http://proteopedia.org/wiki/index.php/G_protein-coupled_receptor#3D_Structures_of_G_protein-coupled_receptors G-protein coupled receptor]<ref>PMID:23237917</ref>. Its structure is similar to the [[Metabotropic glutamate receptor|metabotropic glutamate receptors]] (mGluR) ligand binding domain. The GABA<sub>B</sub> receptor is central to inhibitory neurotransmission in the brain and therefor considered as a good candidate for treatments against alcoholism, stress and some brain diseases<ref>PMID:19913201</ref>. | |
| - | + | The GABA<sub>B</sub> receptor can stimulate the opening of the K<sup>+</sup> (Potassium) channels in the postsynaptic membrane, bringing the neuron closer to the [https://www.youtube.com/watch?v=4kx9_0YwShE equilibrium potential] of K<sup>+</sup>, producing [http://en.wikipedia.org/wiki/Hyperpolarization_(biology) hyperpolarization]. As a result, the Ca<sup>+2</sup> (Calcium) channels in the presynaptic terminal close, and neurotransmitter release stops. In addition GABA<sub>B</sub> receptor function lead to the reduction adenylyl cyclase's activity and decrease the cell’s conductance to Ca<sup>+2</sup> .[http://physrev.physiology.org/content/84/3/835.short]. | |
| - | + | [[Image:GABAb.receptor.cartoon2.png|250px|thumb|GABAb receptors are formed from the heterodimerization of two similar 7TM subunits termed GABAB1 and GABAB2 ]] | |
| - | The GABA<sub>B</sub> receptor | + | =='''''Structure'''''== |
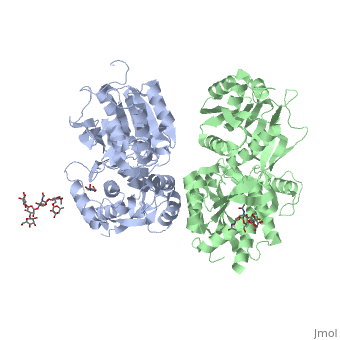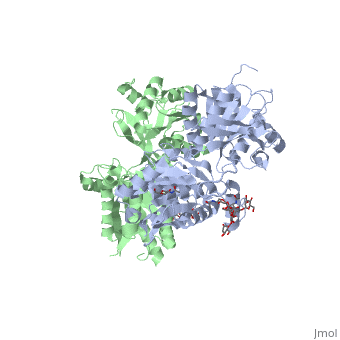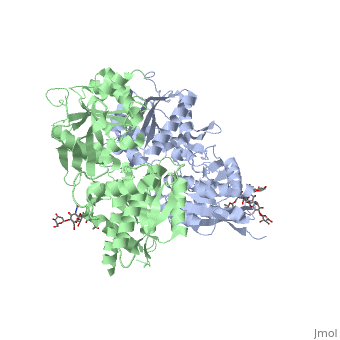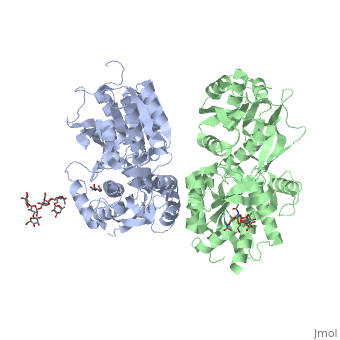
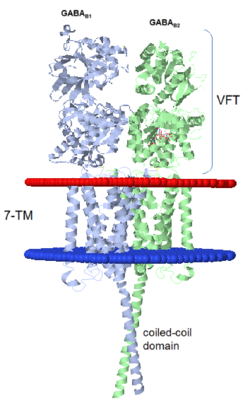
| - | ==''''' | + | GABA<sub>B</sub> receptor functions as an obligatory heterodimer subunit of '''GABA<sub>B1</sub>''' (GBR1) and '''GABA<sub>B2</sub>''' (GBR2). '''GBR1''' (blue) is responsible for ligand-binding. '''GBR2''' (green), on the other hand, is responsible for G protein coupling subunit. The GABA<sub>B</sub> receptor is one of the few obligate receptor heterodimer currently known. There is no crystal or NMR structure of the complete receptor since it has extracellular and inter cellular regions. |
| - | + | Each subunit, GBR1 and GBR2, is a domain of seven-transmembrane helices, composed of a large extracellular domain called [http://en.wikipedia.org/wiki/Venus_flytrap venus flytrap], and intercellular domain which is important for the dimerzation. | |
| - | + | ==='''The extracellular domain VFT'''=== | |
| - | + | The VFT contains two lobe-shaped domains: LB1 and LB2, which are connected by three short loops. | |
| - | + | ||
LB1 and LB2 are <scene name='70/701448/Gbr2_subunit/1'>αβ-folds composed of central β sheet flanked by an α helix</scene><ref>PMID:24305054</ref>. | LB1 and LB2 are <scene name='70/701448/Gbr2_subunit/1'>αβ-folds composed of central β sheet flanked by an α helix</scene><ref>PMID:24305054</ref>. | ||
| - | <Structure load='4mqe' size='350' frame='true' align='right' caption='Insert caption here' scene='Insert optional scene name here' /> | ||
| - | + | '''Common subunit-subunit interactions''' | |
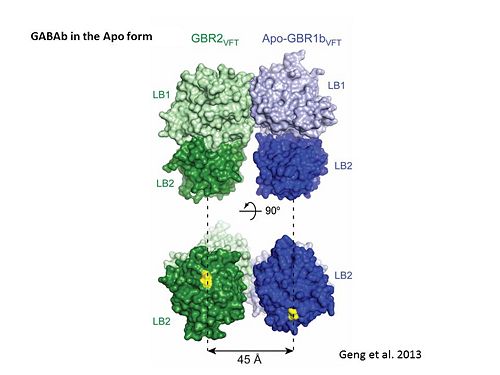
| - | + | In both the resting (apo form, PDB [[4mqe|4MQE]]) and active states (the GABA<sub>B</sub> agonist-bound structures), GBR1bVFT and GBR2VFT interact through their LB1 domains. In the apo and antagonist-bound (inactive state) structures, the subunit association is exclusively facilitated by this LB1-LB1 contact. The heterodimer buries over 1,400 Å2 of solvent accessible surface area and exhibits exceptionally high interfacial shape correlation. <scene name='70/701448/Lb1_lb1_interaction/1'>The LB1-LB1 interaction is mediated by the B and C helices of both subunits.</scene> | |
| - | + | ===='''''Agonist and antagonist binding'''''==== | |
| + | All of the [http://en.wikipedia.org/wiki/Agonist agonists] and [http://en.wikipedia.org/wiki/Receptor_antagonist antagonists] bind the '''extracellular VFT module situated at the crevice between the LB1 and LB2 domains of the GBR1 subunit'''. | ||
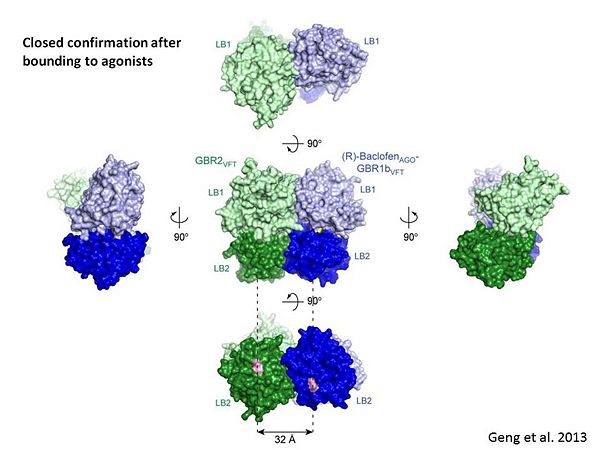
| - | + | The agonist binding induces the formation of an additional heterodimer interface between the LB2 domains of GBR1bVFT and GBR2VFT subunits. The LB2-LB2 interface buries over 1,300 Å2 of solvent accessible surface area, has poor shape complementarity, and is dominated by polar interactions. The LB2-LB2 interaction is mediated by two strand-loop-helix motifs from each LB2 domain. <scene name='70/701448/Lb2-lb2_interaction/3'> The neighboring strands f and g are part of the central β-sheet in LB2, and helices F and G flank the β-sheet</scene>. (GBR1:LB2 :- strand f-red, strand g-blue, helix F-orange, helix G-darkochid. GBR2:LB2 :- strand f-dimgray, strand g-yellow, helix F-hotpink, helix G-brown). | |
| - | + | LB1 and LB2 resdues of GBR1 interact with the GABA<sub>B</sub> agonsits such as: [http://en.wikipedia.org/wiki/Gamma-Aminobutyric_acid GABA],[http://en.wikipedia.org/wiki/Baclofen baclofen]. as a results of this interacting '''closed conformation''' will be stabilized when <scene name='70/701448/Gaba_ligand/4'> GBR1 subunit bound to GABA</scene> (PDB [[4ms3|4MS3]]) or <scene name='70/701448/Baclofen/3'>bound to baclofen</scene> (PDB [[4ms4|4MS4]]) and other agonists. | |
| - | [[Image: | + | [[Image:GABAb receptor in the Apo form.jpg|center|500px|[[4mqe]]]] |
| - | + | [[Image:GABAb bound to agonist.jpg|center|thumb|600px|Agonist-induced conformational changes, the substantial rearrangement of the LB2 domains from the apo to the agonist-bound state shortens the distance between the C-termini of the two subunits from 45 Å to 32 Å]] | |
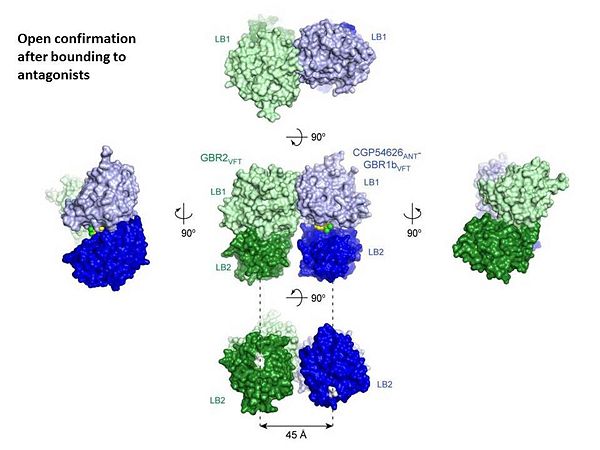
| + | GABA<sub>B</sub> antgonists such as: [http://en.wikipedia.org/wiki/Saclofen saclofen], [http://www.hellobio.com/cgp-46381.html CGP46381], [http://www.hellobio.com/phaclofen.html?picat=&q=phaclofen phaclofen], [http://www.hellobio.com/cgp-35348.html?picat=&q=CGP+35348 CGP35348] and [http://www.hellobio.com/cgp-54626-hydrochloride.html?picat=&q=CGP+54626 CGP54626], mostly intercat with LB1 resduses of GBR1. both ends of the antagonist bind the LB1, which produce '''open confirmation''' of GBR1. | ||
| + | it is very easy to notice the '''open confirmation''' when : | ||
| + | <scene name='70/701448/Saclofen_ant/2'>GBR1 subunit bound to saclofen</scene> (PDB [[4mqf|4MQF]]). | ||
| + | <scene name='70/701448/Cgp46381_ant/2'>GBR1 subunit bound to CGP46381</scene> (PDB [[4ms1|4MS1]]). | ||
| + | <scene name='70/701448/Phaclofen_ant/1'>GBR1 subunit bound to phaclofen</scene> (PDB [[4mrm|4MRM]]). | ||
| + | <scene name='70/701448/Ant_cg835348/1'>GBR1 subunit bound to CGP35348</scene> (PDB [[4mr8|4MR8]]). | ||
| + | <scene name='70/701448/Ant_CGP54626/1'>GBR1 subunit bound to CGP54626</scene> (PDB [[4mr7|4MR7]]). | ||
| + | [[Image:GABAb.bound.to.antagonist.jpg|center|thumb|600px|The ligand-binding cleft of GBR1bVFT stays open with each bound antagonist. In addition, GBR2VFT remains wide open with an empty interdomain cleft. This open-open configuration of the apo and antagonist-bound structures corresponds to the | ||
| + | resting (or inactive) state of the heterodimeric receptor (Geng et al 2013).]] | ||
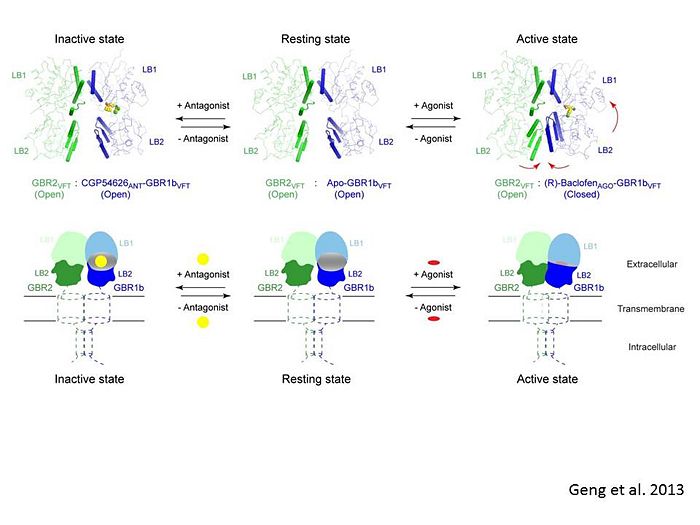
| - | + | In short, the apo and antagonist-bound structures represent the resting state of the receptor; the agonist-bound complex corresponds to the | |
| - | + | active state. Both subunits, GBR1 and GBR2 adopt an open conformation at rest, and only GBR1 closes upon agonist-induced receptor activation. | |
| + | [[Image:Summery for the mechanism.jpg|center|thumb|700px|Schematic diagram showing the conformational equilibrium of full length GABAb receptor (Geng et al 2013)]] | ||
| - | === | + | ==='''''The intercellular dimerization motif'''''=== |
| + | When the GBR1 subunit is expressed alone, it is trapped in vesicles within the cell, whereas the GBR2 alone is expressed on the cell surface, but cannot bind GABA or activate G proteins. When both receptor subunits are expressed in the same cell, the receptor interact through <scene name='70/701448/C_termenal_coild_coil_domain/1'>coiled-coil domain</scene><ref>PMID:24778228</ref><ref>PMID:12209124</ref> (PDB [[4pas|4PAS]]) in their carboxyl tails. Then they are expressed on the cell surface, bind GABA and activate G proteins. | ||
| + | This domain is shaped by <scene name='70/701448/Coild-coil_domain/1'>polar interactions within the hydrophobic core</scene> and is stabilized by <scene name='70/701448/C_termenal_coild_coil_domain/3'>hydrogen-bonding interactions</scene><ref>PMID:24778228</ref>. | ||
| - | + | == '''''Mutagenesis studies of GBR1 and GBR2 in the extracellular domain VFT''''' == | |
| - | + | '''Mutagenesis studies in GBR1 subunit''' [http://www.uniprot.org/uniprot/Q9UBS5#pathology_and_biotech] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <scene name='70/701448/Gbr14mqe/2'>Trp182Ala</scene>, <scene name='70/701448/Gbr14mqe/4'>Trp395Ala</scene>, : Abolishes signaling via G-proteins and antagonist binding. | |
| - | + | <scene name='70/701448/Gbr14mqe/6'>Tyr230Ala</scene>: Slightly decreases signaling via G-proteins. | |
| - | + | ||
| - | + | ||
| - | + | <scene name='70/701448/Gbr14mqe/7'>Tyr234Ala</scene>: Decreases signaling via G-proteins. | |
| - | <scene name='70/701448/ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | = | + | <scene name='70/701448/Gbr14mqe/8'>His287Ala</scene>: Strongly reduces signaling via G-proteins and abolishes antagonist binding. |
| - | + | <scene name='70/701448/Gbr14mqe/10'>Tyr367Ala</scene>: Strongly reduces signaling via G-proteins, no effect on antagonist binding. | |
| + | '''Mutagenesis studies in GBR2 subunit''' [http://www.uniprot.org/uniprot/O75899#pathology_and_biotech] | ||
| + | |||
| + | <scene name='70/701448/4mqegbr2/1'>Tyr118Ala</scene>: Decreases signaling via G-proteins. | ||
== References == | == References == | ||
<references/> | <references/> | ||
| - | [[Link title]] | ||
Current revision
| |||||||||||