Adrenodoxin reductase
From Proteopedia
(Difference between revisions)
(Introduction) |
|||
| (29 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
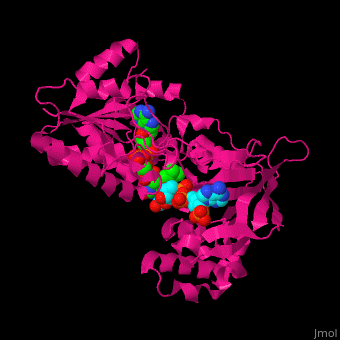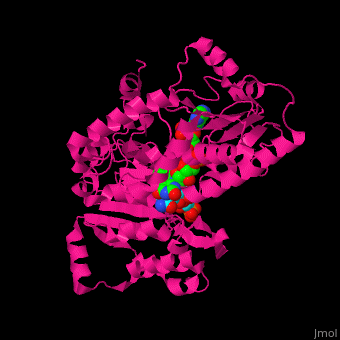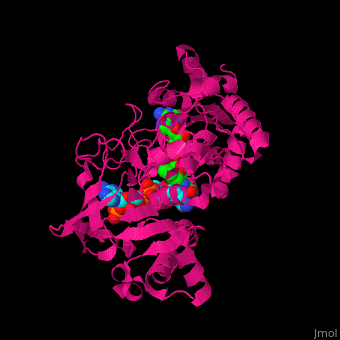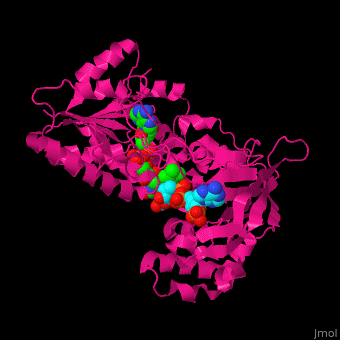
| - | Adrenodoxin reductase (AR) is | + | <StructureSection load='' size='450' side='right' scene='70/702915/Cv/1' caption='Structure of adrenodoxin reductase with FAD and NADP. PDB ID: [[1e1k]].'> |
| + | __TOC__ | ||
| + | ==Function== | ||
| + | |||
| + | '''Adrenodoxin reductase''' (AR) is an FAD containing flavoprotein that functions as an electron transfer protein in the mitochondrial P450 systems that catalyze essential steps in the biosynthesis of steroid hormones.<ref name="HI-review">PMID:22217824</ref> | ||
| + | |||
| + | The enzyme nomenclature code of AR is [http://www.chem.qmul.ac.uk/iubmb/enzyme/EC1/18/1/6.html 1.18.1.6]. | ||
| + | |||
| + | ==Mitochondrial P450 systems== | ||
| + | Mitochondrial P450 systems are expressed in cells that specialize in the biosynthesis and secretion of steroids. The reactions these systems catalyze include the first step in steroid hormone synthesis that is cleavage of the side chain of cholesterol producing pregnenolone, C11- hydroxylation of steroids producing glucocorticoids, C-18 hydroxylation of steroids leading to the synthesis of mineralocorticoids, and C-25 and C-27 hydroxylations of steroids in the pathways of bile acid and vitamin D synthesis.<ref name="HI-review" /> | ||
| + | |||
| + | Mitochondrial P450 systems are composed of three proteins: | ||
| + | * Adrenodoxin reductase | ||
| + | * Adrenodoxin, a [2Fe-2S] ferredoxin type iron-sulfur protein | ||
| + | * Mitochondrial P450 | ||
| + | |||
| + | The first two electron transfer proteins are shared by all systems. Substrate and reaction specificity of the system is dependent on the type of P450. | ||
| + | |||
| + | The reactions catalyzed by P450 type enzymes are called monooxygenation reactions because P450 catalyzes incorporation of only one atom of molecular O<sub>2</sub> into substrate (S) while reducing the second atom of O into H<sub>2</sub>O with the following stoichiometry: | ||
| + | |||
| + | SH + O<sub>2</sub> + NAD(P)H + H<sup>+</sup> → ROH + H<sub>2</sub>O + NAD(P)<sup>+</sup> | ||
| + | |||
| + | In the mitochondrial P450 systems, the source of electrons is NADPH. The main function of AR in these systems is to accept two electrons from NADPH which are transferred to FAD with the following stoichiometry: | ||
| + | |||
| + | NADPH + H<sup>+</sup> + FAD → NADP<sup>+</sup> + FADH<sub>2</sub> | ||
| + | |||
| + | After this initial transfer, AR catalyzes transfer of electron from FADH<sub>2</sub> to adrenodoxin in two independent steps as adrenodoxin is a single electron acceptor. Reduced adrenodoxin in turn transfers its electron to mitochondrial P450 during the complex catalytic cycle of P450. | ||
| + | |||
| + | Thus, the route of electron transfer in these systems is as follows: | ||
| + | |||
| + | NADPH → AR → adrenodoxin → P450 | ||
| + | |||
| + | ==Structure of adrenodoxin reductase== | ||
| + | |||
| + | AR has two main domains. The N terminal domain contains a [[Rossmann fold]] that binds FAD and a central domain that binds NADPH.<ref>PMID:29177972</ref> | ||
| + | |||
| + | The following scenes illustrate some aspects of the structure. | ||
| + | |||
| + | To rotate the molecule click and hold left mouse button. | ||
| + | |||
| + | To zoom-in or zoom-out first click on the structure and then use the mouse wheel. | ||
| + | |||
| + | Click the following green links for the action indicated: | ||
| + | |||
| + | <scene name='70/702915/Cv/14'>Ball and stick repsentation of NADP and FAD in AR</scene> | ||
| + | |||
| + | <scene name='70/702915/Cv/15'>Spacefill repsentation of NADP and FAD in AR</scene> | ||
| + | |||
| + | <scene name='70/702915/Cv/16'>FAD binding site in AR</scene> (water molecules shown as red spheres) | ||
| + | |||
| + | <scene name='70/702915/Cv/17'>NADP binding site in AR</scene> (water molecules shown as red spheres) (PDB entry [[1e1k]])<ref>PMID:10998235</ref> | ||
| + | |||
| + | <scene name='70/702915/Fad_rossmann_fold/2'>Rossmann fold of FAD</scene> | ||
| + | {{clear}} | ||
| + | |||
| + | ==3D Structures of Adrenodoxin reductase== | ||
| + | [[Adrenodoxin reductase 3D structures]] | ||
| + | |||
| + | </StructureSection> | ||
==References== | ==References== | ||
<references /> | <references /> | ||
| - | + | [[Category: Oxidoreductase]] | |
| - | [[ | + | [[Category: Rossmann fold]] |
| + | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
References
- ↑ 1.0 1.1 Hanukoglu I. Steroidogenic enzymes: structure, function, and role in regulation of steroid hormone biosynthesis. J Steroid Biochem Mol Biol. 1992 Dec;43(8):779-804. doi:, 10.1016/0960-0760(92)90307-5. PMID:22217824 doi:http://dx.doi.org/10.1016/0960-0760(92)90307-5
- ↑ Hanukoglu I. Conservation of the Enzyme-Coenzyme Interfaces in FAD and NADP Binding Adrenodoxin Reductase-A Ubiquitous Enzyme. J Mol Evol. 2017 Dec;85(5-6):205-218. doi: 10.1007/s00239-017-9821-9. Epub 2017, Nov 24. PMID:29177972 doi:http://dx.doi.org/10.1007/s00239-017-9821-9
- ↑ Ziegler GA, Schulz GE. Crystal structures of adrenodoxin reductase in complex with NADP+ and NADPH suggesting a mechanism for the electron transfer of an enzyme family. Biochemistry. 2000 Sep 12;39(36):10986-95. PMID:10998235
Proteopedia Page Contributors and Editors (what is this?)
Alexander Berchansky, Israel Hanukoglu, Michal Harel, Joel L. Sussman