FMDV 3C
From Proteopedia
| (12 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
FMDV 3C Protease | FMDV 3C Protease | ||
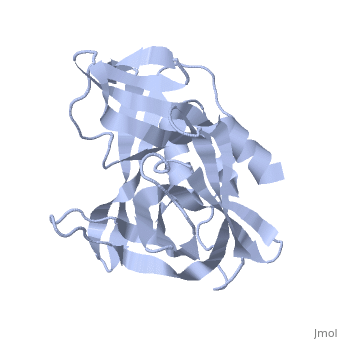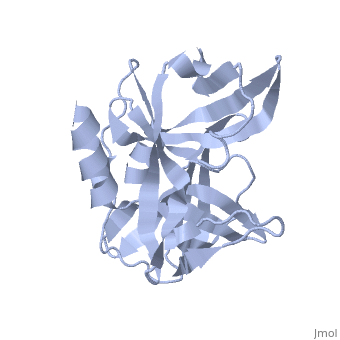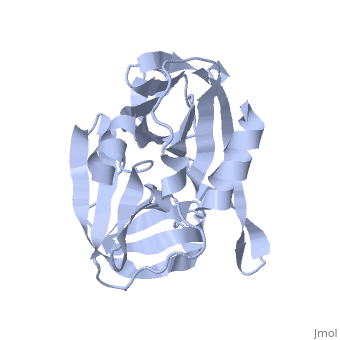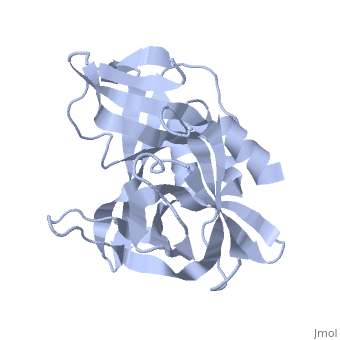
| - | <StructureSection load='2j92' size='340' side='right' caption='FMDV 3C Protease Type A10' scene=''> | + | <StructureSection load='2j92' size='340' side='right' caption='FMDV 3C Protease Type A10 (PDB code [[2j92]])' scene=''> |
| - | + | '''Foot and Mouth Disease 3C Protease'''. | |
| - | The viral | + | '''By, Erica Martel''' |
| - | [[Image:FMDV_3C_Prot.jpeg]] | + | |
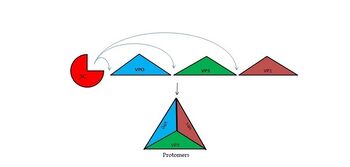
| + | The FMDV viral genome is translated as a single polypeptide precursor that must be cleaved into functional proteins by virally encoded proteases. The FMDV 3C protease is responsible for most cleavages of the viral polyprotein (10 out of the 13 cleavages) and has a highly conserved amino acid sequence across different viral serotypes, making the enzyme an attractive target for inhibitor design and antiviral drugs. The 3C protease cuts itself out of the polyprotein and is responsible for processing the P1 peptide into three viral peptides, VP0 VP3, and VP1. The VP0 peptide is further processed into VP4 and VP2 by an unknown mechanism. To form the FMDV capsid fully processed VPs assemble into a triangular protomer, Figure below. Five protomers come together to make a pentamer, twelve pentamers conjoin to complete the virus shell. | ||
| + | |||
| + | |||
| + | [[Image:FMDV_3C_Prot.jpeg|350px|left|thumb]] | ||
== Function == | == Function == | ||
| - | FMDV 3C has multiple functions which will be discussed below: | ||
| - | + | The 3C only known function of the viral 3C protease was processing of the viral polyprotein, until recent research discovered interactions with host cell proteins. The 3C protease has been shown to induce proteolytic cleavage of Histone H3. As well as rapid inhibition of host cell protein synthesis by cleavage of translation initiation factors eIF4A, eIF4G between residues E143 and V144. | |
| + | 3C is also able to induce Golgi Fragmentation, causing fragmentation of early, medial, and late Golgi compartments with the most pronounced effect being on early Golgi compartments. Research has found that the Golgi fragments were still able to receive membrane proteins from the endoplasmic reticulum however they were unable to transfer protein to the plasma membrane. | ||
| + | 3C has been shown to be able to cleave the C terminus of the nuclear RNA binding protein SAM 68 resulting in redistribution to the cytoplasm. This was particularly interesting because of the fact that Sam68 has been shown to effect FMDV post entry events by interacting with the internal ribosomal entry site within the 5′ non-translated region of the FMDV genome, and Sam68 knockdown decreased FMDV IRES-driven activity in vitro suggesting that it could modulate translation of the viral genome. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | == Disease == | ||
| + | FMDV, the etiologic agent of Foot-and-Mouth Disease (FMD), affects a variety of cloven-hoofed animals in different parts of the world, but to date is not present in North America or Australia. FMDV is extremely infectious and displays high adaptability as well as short incubation times. This virus exists in the form of seven different serotypes (A, O, C, Asia-1, and South African Territories - SAT - 1, 2, and 3) and it has innumerable subtypes. These factors, in conjunction with ecological dynamics of livestock, result in a rapid, widespread, and devastating disease that causes exorbitant economic losses for the agricultural industry. FMDV outbreaks impact local and global food-animal markets as trade regulations restrict exportation of products from FMDV endemic areas, stressing the need for a reliable diagnostic tool and effective, yet safe, vaccinations. FMDV is also on the “A” list of infectious diseases of animals of the OIE, the world organization for animal health which means that it is a transmissible disease that has the potential for very rapid spread and is of serious socioeconomic importance. | ||
== Relevance == | == Relevance == | ||
| - | Vaccines against FMDV are used where the disease is endemic | + | |
| - | + | Vaccines against FMDV exist and are used where the disease is endemic. The primary vaccine utilized in endemic areas is inactivated FMDV. The inactive FMDV vaccine has drawbacks such as difficulty telling vaccinated from infected animals and it carries the risk that a FMDV outbreak will be caused by incomplete inactivation of the vaccine virus. As a result, there has been interest in producing empty virus like particles (VLPs) for use as vaccines against FMD. Due to the FMDV 3C protease activity on host cell proteins producing large amounts of VLPs has been difficult. | |
| - | + | ||
== Structural highlights == | == Structural highlights == | ||
<scene name='70/702408/Fmdv_3c_protease_type_a10/6'>FMDV 3C Protease</scene> | <scene name='70/702408/Fmdv_3c_protease_type_a10/6'>FMDV 3C Protease</scene> | ||
| - | Click " <scene name="/12/3456/Sample/1">color</scene> " to view by Group, | + | Click " <scene name="/12/3456/Sample/1">color</scene> " to view by Group, or <scene name="/12/3456/Sample/2"> shape </scene> to make transparent representation of the protein. |
</StructureSection> | </StructureSection> | ||
| + | |||
| + | |||
== References == | == References == | ||
| + | Klump, W., O. Marquardt, and P.H. Hofschneider, Biologically active protease of foot and mouth disease virus is expressed from cloned viral cDNA in Escherichia coli. Proc Natl Acad Sci U S A, 1984. 81(11): p. 3351-5. | ||
| + | |||
| + | Grubman, M.J., M. Zellner, and J. Wagner, Antigenic comparison of the polypeptides of foot-and-mouth disease virus serotypes and other picornaviruses. Virology, 1987. 158(1): p. 133-40. | ||
| + | |||
| + | Vakharia, V.N., et al., Proteolytic processing of foot-and-mouth disease virus polyproteins expressed in a cell-free system from clone-derived transcripts. J Virol, 1987. 61(10): p. 3199-207. | ||
| + | |||
| + | Grubman, M.J. and B. Baxt, Translation of foot-and-mouth disease virion RNA and processing of the primary cleavage products in a rabbit reticulocyte lysate. Virology, 1982. 116(1): p. 19-30. | ||
| + | |||
| + | 5.ablanian, G.M. and M.J. Grubman, Characterization of the foot-and-mouth disease virus 3C protease expressed in Escherichia coli. Virology, 1993. 197(1): p. 320-7. | ||
| + | |||
| + | Ryan, M.D., G.J. Belsham, and A.M. King, Specificity of enzyme-substrate interactions in foot-and-mouth disease virus polyprotein processing. Virology, 1989. 173(1): p. 35-45. | ||
| + | |||
| + | Clarke, B.E. and D.V. Sangar, Processing and assembly of foot-and-mouth disease virus proteins using subgenomic RNA. J Gen Virol, 1988. 69 ( Pt 9): p. 2313-25. | ||
| + | |||
| + | Grubman, M.J., et al., Identification of the active-site residues of the 3C proteinase of foot-and-mouth disease virus. Virology, 1995. 213(2): p. 581-9. | ||
| + | |||
| + | Falk, M.M., et al., Foot-and-mouth disease virus protease 3C induces specific proteolytic cleavage of host cell histone H3. J Virol, 1990. 64(2): p. 748-56. | ||
| + | |||
| + | Belsham, G.J., G.M. McInerney, and N. Ross-Smith, Foot-and-mouth disease virus 3C protease induces cleavage of translation initiation factors eIF4A and eIF4G within infected cells. J Virol, 2000. 74(1): p. 272-80. | ||
| + | |||
| + | Strong, R. and G.J. Belsham, Sequential modification of translation initiation factor eIF4GI by two different foot-and-mouth disease virus proteases within infected baby hamster kidney cells: identification of the 3Cpro cleavage site. J Gen Virol, 2004. 85(Pt 10): p. 2953-62. | ||
| + | |||
| + | Devaney, M.A., et al., Leader protein of foot-and-mouth disease virus is required for cleavage of the p220 component of the cap-binding protein complex. J Virol, 1988. 62(11): p. 4407-9. | ||
| + | |||
| + | Medina, M., et al., The two species of the foot-and-mouth disease virus leader protein, expressed individually, exhibit the same activities. Virology, 1993. 194(1): p. 355-9. | ||
| + | |||
| + | Li, W., et al., Cleavage of translation initiation factor 4AI (eIF4AI) but not eIF4AII by foot-and-mouth disease virus 3C protease: identification of the eIF4AI cleavage site. FEBS Lett, 2001. 507(1): p. 1-5. | ||
| + | |||
| + | Zhou, Z., et al., Foot-and-mouth disease virus 3C protease induces fragmentation of the Golgi compartment and blocks intra-Golgi transport. J Virol, 2013. 87(21): p. 11721-9. | ||
| + | |||
| + | Armer, H., et al., Foot-and-mouth disease virus, but not bovine enterovirus, targets the host cell cytoskeleton via the nonstructural protein 3Cpro. J Virol, 2008. 82(21): p. 10556-66. | ||
| + | |||
| + | Lawrence, P., E.A. Schafer, and E. Rieder, The nuclear protein Sam68 is cleaved by the FMDV 3C protease redistributing Sam68 to the cytoplasm during FMDV infection of host cells. Virology, 2012. 425(1): p. 40-52. | ||
| + | |||
| + | Wang, D., et al., Foot-and-mouth disease virus 3C protease cleaves NEMO to impair innate immune signaling. J Virol, 2012. 86(17): p. 9311-22. | ||
| + | |||
| + | Martinez-Salas, E. and E. Domingo, Effect of expression of the aphthovirus protease 3C on viral infection and gene expression. Virology, 1995. 212(1): p. 111-20. | ||
| + | |||
| + | Birtley, J.R., et al., Crystal structure of foot-and-mouth disease virus 3C protease. New insights into catalytic mechanism and cleavage specificity. J Biol Chem, 2005. 280(12): p. 11520-7. | ||
| + | |||
| + | Sweeney, T.R., et al., Structural and mutagenic analysis of foot-and-mouth disease virus 3C protease reveals the role of the beta-ribbon in proteolysis. J Virol, 2007. 81(1): p. 115-24. | ||
| + | |||
| + | |||
<references/> | <references/> | ||
references <ref>DOI 10.1002/ijch.201300024</ref> | references <ref>DOI 10.1002/ijch.201300024</ref> | ||
Current revision
FMDV 3C Protease
| |||||||||||
References
Klump, W., O. Marquardt, and P.H. Hofschneider, Biologically active protease of foot and mouth disease virus is expressed from cloned viral cDNA in Escherichia coli. Proc Natl Acad Sci U S A, 1984. 81(11): p. 3351-5.
Grubman, M.J., M. Zellner, and J. Wagner, Antigenic comparison of the polypeptides of foot-and-mouth disease virus serotypes and other picornaviruses. Virology, 1987. 158(1): p. 133-40.
Vakharia, V.N., et al., Proteolytic processing of foot-and-mouth disease virus polyproteins expressed in a cell-free system from clone-derived transcripts. J Virol, 1987. 61(10): p. 3199-207.
Grubman, M.J. and B. Baxt, Translation of foot-and-mouth disease virion RNA and processing of the primary cleavage products in a rabbit reticulocyte lysate. Virology, 1982. 116(1): p. 19-30.
5.ablanian, G.M. and M.J. Grubman, Characterization of the foot-and-mouth disease virus 3C protease expressed in Escherichia coli. Virology, 1993. 197(1): p. 320-7.
Ryan, M.D., G.J. Belsham, and A.M. King, Specificity of enzyme-substrate interactions in foot-and-mouth disease virus polyprotein processing. Virology, 1989. 173(1): p. 35-45.
Clarke, B.E. and D.V. Sangar, Processing and assembly of foot-and-mouth disease virus proteins using subgenomic RNA. J Gen Virol, 1988. 69 ( Pt 9): p. 2313-25.
Grubman, M.J., et al., Identification of the active-site residues of the 3C proteinase of foot-and-mouth disease virus. Virology, 1995. 213(2): p. 581-9.
Falk, M.M., et al., Foot-and-mouth disease virus protease 3C induces specific proteolytic cleavage of host cell histone H3. J Virol, 1990. 64(2): p. 748-56.
Belsham, G.J., G.M. McInerney, and N. Ross-Smith, Foot-and-mouth disease virus 3C protease induces cleavage of translation initiation factors eIF4A and eIF4G within infected cells. J Virol, 2000. 74(1): p. 272-80.
Strong, R. and G.J. Belsham, Sequential modification of translation initiation factor eIF4GI by two different foot-and-mouth disease virus proteases within infected baby hamster kidney cells: identification of the 3Cpro cleavage site. J Gen Virol, 2004. 85(Pt 10): p. 2953-62.
Devaney, M.A., et al., Leader protein of foot-and-mouth disease virus is required for cleavage of the p220 component of the cap-binding protein complex. J Virol, 1988. 62(11): p. 4407-9.
Medina, M., et al., The two species of the foot-and-mouth disease virus leader protein, expressed individually, exhibit the same activities. Virology, 1993. 194(1): p. 355-9.
Li, W., et al., Cleavage of translation initiation factor 4AI (eIF4AI) but not eIF4AII by foot-and-mouth disease virus 3C protease: identification of the eIF4AI cleavage site. FEBS Lett, 2001. 507(1): p. 1-5.
Zhou, Z., et al., Foot-and-mouth disease virus 3C protease induces fragmentation of the Golgi compartment and blocks intra-Golgi transport. J Virol, 2013. 87(21): p. 11721-9.
Armer, H., et al., Foot-and-mouth disease virus, but not bovine enterovirus, targets the host cell cytoskeleton via the nonstructural protein 3Cpro. J Virol, 2008. 82(21): p. 10556-66.
Lawrence, P., E.A. Schafer, and E. Rieder, The nuclear protein Sam68 is cleaved by the FMDV 3C protease redistributing Sam68 to the cytoplasm during FMDV infection of host cells. Virology, 2012. 425(1): p. 40-52.
Wang, D., et al., Foot-and-mouth disease virus 3C protease cleaves NEMO to impair innate immune signaling. J Virol, 2012. 86(17): p. 9311-22.
Martinez-Salas, E. and E. Domingo, Effect of expression of the aphthovirus protease 3C on viral infection and gene expression. Virology, 1995. 212(1): p. 111-20.
Birtley, J.R., et al., Crystal structure of foot-and-mouth disease virus 3C protease. New insights into catalytic mechanism and cleavage specificity. J Biol Chem, 2005. 280(12): p. 11520-7.
Sweeney, T.R., et al., Structural and mutagenic analysis of foot-and-mouth disease virus 3C protease reveals the role of the beta-ribbon in proteolysis. J Virol, 2007. 81(1): p. 115-24.
references [1]