Regulator of G protein signaling
From Proteopedia
(Difference between revisions)
| (18 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
==Regulator of G protein signaling (RGS) interactions with G proteins – RGS4-Gα<sub>i</sub> as a model structure.== | ==Regulator of G protein signaling (RGS) interactions with G proteins – RGS4-Gα<sub>i</sub> as a model structure.== | ||
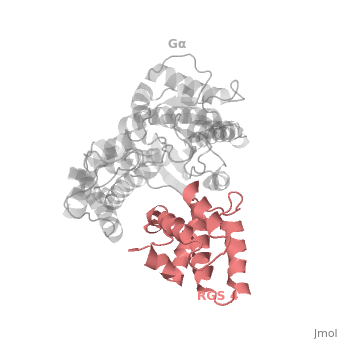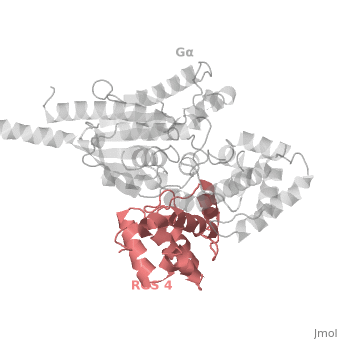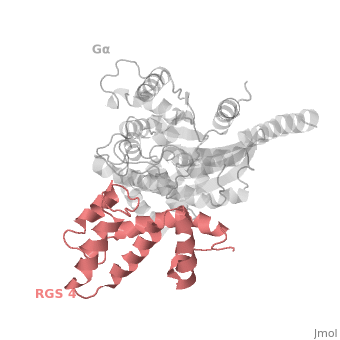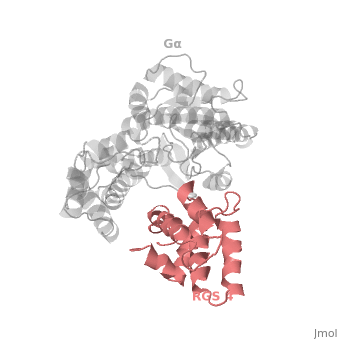
| - | <StructureSection load=' | + | <StructureSection load='' size='340' side='right' caption='Rat regulator of G-protein signalling 4 (red) complex with guanine nucleotide-binding protein α subunit (grey) (PDB code [[1agr]])' scene='70/701447/Gi-rgs4/9' pspeed='8'> |
== Heterotrimeric G-proteins family == | == Heterotrimeric G-proteins family == | ||
Human heterotrimeric G-proteins are derived from 35 genes: 16 encoding α subunits, 5 β and 14 γ subunits. | Human heterotrimeric G-proteins are derived from 35 genes: 16 encoding α subunits, 5 β and 14 γ subunits. | ||
| Line 6: | Line 6: | ||
G-proteins interact with diverse protein partners, such as G-protein coupled receptors (GPCRs), downstream effectors, and other proteins. | G-proteins interact with diverse protein partners, such as G-protein coupled receptors (GPCRs), downstream effectors, and other proteins. | ||
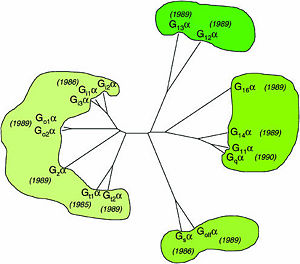
One important G-protein interaction is with members of the RGS protein family. This interaction occurs when the G-protein alpha subunit is activated, and depends on the Gα class, which in turn depends on their sequence that classifies them into several sub-types. Based on the Phylogenetic tree of mammalian G-protein, G-protein α-subunits are classified to 4 groups based on their sequence identity. The first two groups, Gα<sub>i</sub> and Gα<sub>q</sub> families have selectivity towards The RGS members of R4-subfamily and R12-subfamily. Another group, Gα<sub>z</sub> subunits were suggested to have selectivity for the RGS members of RZ-subfamily. The last subfamilies Gα<sub>s</sub> and Gα<sub>12/13</sub> might interact with diverse proteins subfamilies that include the ~120-residue RGS homology domain but can't interact with canonical RGS members. | One important G-protein interaction is with members of the RGS protein family. This interaction occurs when the G-protein alpha subunit is activated, and depends on the Gα class, which in turn depends on their sequence that classifies them into several sub-types. Based on the Phylogenetic tree of mammalian G-protein, G-protein α-subunits are classified to 4 groups based on their sequence identity. The first two groups, Gα<sub>i</sub> and Gα<sub>q</sub> families have selectivity towards The RGS members of R4-subfamily and R12-subfamily. Another group, Gα<sub>z</sub> subunits were suggested to have selectivity for the RGS members of RZ-subfamily. The last subfamilies Gα<sub>s</sub> and Gα<sub>12/13</sub> might interact with diverse proteins subfamilies that include the ~120-residue RGS homology domain but can't interact with canonical RGS members. | ||
| - | + | ||
| - | Image:Ga_family_figure1-heterotrimeric_G-protein-short_history_06.jpg|300px|Homology of mammalian G-protein α-subunits. | + | [[Image:Ga_family_figure1-heterotrimeric_G-protein-short_history_06.jpg|300px|Homology of mammalian G-protein α-subunits.]] |
| - | + | ||
| - | + | ||
Phylogenetic tree of mammalian G-protein α-subunits classified to 4 groups based on their sequence identity.<ref name="Milligan2006">PMID: 16402120</ref> | Phylogenetic tree of mammalian G-protein α-subunits classified to 4 groups based on their sequence identity.<ref name="Milligan2006">PMID: 16402120</ref> | ||
== RGS proteins == | == RGS proteins == | ||
| - | Regulator of G-proteins signaling (RGS) proteins play a critical role in many G protein-dependent signaling pathways. Thus, RGS proteins have been implicated in a wide range of pathologies, including cancer, hypertension, arrhythmias, drug abuse and schizophrenia. RGS proteins ‘turn off’ heterotrimeric (αβγ) G-proteins and thereby determine the duration of G protein–mediated signaling events. Therefore, RGS proteins function as GTPase Activating Proteins (GAPs), and this GAP activity is mediated by allosteric interactions. | + | '''Regulator of G-proteins signaling''' (RGS) proteins play a critical role in many G protein-dependent signaling pathways. Thus, RGS proteins have been implicated in a wide range of pathologies, including cancer, hypertension, arrhythmias, drug abuse and schizophrenia. RGS proteins ‘turn off’ heterotrimeric (αβγ) G-proteins and thereby determine the duration of G protein–mediated signaling events. Therefore, RGS proteins function as GTPase Activating Proteins (GAPs), and this GAP activity is mediated by allosteric interactions. |
RGS proteins are selective for binding to the transition state of Gα(GTP → GDP + P<sub>i</sub>). | RGS proteins are selective for binding to the transition state of Gα(GTP → GDP + P<sub>i</sub>). | ||
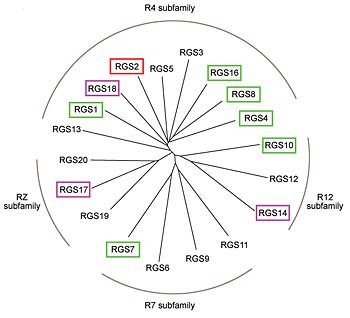
Like many signaling proteins, RGS proteins comprise a large and diverse family. In human genome, Thirty-seven RGS proteins are encoded by gene loci; this collection of related proteins has been divided into 10 different subfamilies based on the relatedness of their RGS domain sequence and their multiple domain architectures. The major subfamily contains About 20 ‘canonical’ RGS proteins that can in theory downregulate any of the 16 activated Gα subunits, although in practice they interact only with members of the G<sub>i</sub> and G<sub>q</sub> families. In these proteins, the ~120-residue RGS homology domain functions as a GTPase-activating protein (GAP) for GTP-bound Gα subunits. In addition to these domains, diverse proteins subfamilies include additional protein-protein interaction domains beyond their signature RGS domain with Gα GAP activity. R7-subfamily members share a multi-domain protein architecture composed of DEP and GGL domains on the N-terminal side of the RGS domain. R12-subfamily members possess a tandem repeat of Ras binding domains (RBDs) and a single GoLoco motif. | Like many signaling proteins, RGS proteins comprise a large and diverse family. In human genome, Thirty-seven RGS proteins are encoded by gene loci; this collection of related proteins has been divided into 10 different subfamilies based on the relatedness of their RGS domain sequence and their multiple domain architectures. The major subfamily contains About 20 ‘canonical’ RGS proteins that can in theory downregulate any of the 16 activated Gα subunits, although in practice they interact only with members of the G<sub>i</sub> and G<sub>q</sub> families. In these proteins, the ~120-residue RGS homology domain functions as a GTPase-activating protein (GAP) for GTP-bound Gα subunits. In addition to these domains, diverse proteins subfamilies include additional protein-protein interaction domains beyond their signature RGS domain with Gα GAP activity. R7-subfamily members share a multi-domain protein architecture composed of DEP and GGL domains on the N-terminal side of the RGS domain. R12-subfamily members possess a tandem repeat of Ras binding domains (RBDs) and a single GoLoco motif. | ||
| - | + | [[Image:Kosloff-NSMB2011-Fig1b.jpg|350px|family of canonical RGS proteins]] | |
| - | Image:Kosloff-NSMB2011-Fig1b.jpg|350px|family of canonical RGS proteins | + | |
| - | + | ||
| - | + | ||
Phylogenetic tree of 19 human RGS domains. | Phylogenetic tree of 19 human RGS domains. | ||
RGS proteins whose activity was tested are colored by their GAP activity, RGS proteins with high GAP activity (green), RGS proteins with low but discernible activities (purple) and RGS2 had no measurable activity (red). | RGS proteins whose activity was tested are colored by their GAP activity, RGS proteins with high GAP activity (green), RGS proteins with low but discernible activities (purple) and RGS2 had no measurable activity (red). | ||
<ref name="Mickey2011">PMID: 21685921</ref> | <ref name="Mickey2011">PMID: 21685921</ref> | ||
| + | |||
| + | == Various RGSs and their functions == | ||
| + | |||
| + | *'''RGS1''' selectively regulates gut T cell trafficking and colitis potential<ref>PMID:21795595</ref>.<br /> | ||
| + | *'''RGS2''' regulates airway hyper responsiveness related to asthma<ref>PMID:25368964</ref>.<br /> | ||
| + | *'''RGS3''' is associated with glioma cell motility<ref>PMID:15055445</ref>.<br /> | ||
| + | *'''RGS4''' is associated with schizophrenia<ref>PMID:15274033</ref>.<br /> | ||
| + | *'''RGS5''' controls blood pressure homeostasis<ref>PMID:23303165</ref>.<br /> | ||
| + | *'''RGS6''' promotes anxiety and depression<ref>PMID:24421401</ref>.<br /> | ||
| + | *'''RGS9-1''' mediates G protein function in photoreceptors<ref>PMID:11601986</ref>.<br /> | ||
| + | *'''RGS10''' negatively regulates cardia remodeling<ref>PMID:26573707</ref>.<br /> | ||
| + | *'''RGS12''' regulates osteoclast differentiation<ref>PMID:25909889</ref>.<br /> | ||
| + | *'''RGS14''' is an selective Hras effector <ref>PMID:19319189</ref>.<br /> | ||
| + | *'''RGS16''' inhibits hepatic fatty acid oxidation<ref>PMID:21357625</ref>.<br /> | ||
| + | *'''RGS17''' is a negative modulator of G protein-coupled receptor signaling in human cancers<ref>PMID:26928451</ref>.<br /> | ||
| + | *'''RGS18''' regulates platelet aggregation and thrombosis<ref>PMID:25969426</ref>.<br /> | ||
== RGS4 Structural highlights == | == RGS4 Structural highlights == | ||
| Line 36: | Line 49: | ||
Many RGS protein residues located in the vicinity of the <scene name='70/701447/Rgs4-ga_interface/4'>RGS domain–Gα interface</scene> (RGS protein shown as wheat cartoon and Gα<sub>i1</sub> subunit shown as white surface) contribute to RGS-G proteins interaction. Based on energy calculation and experimental validation of RGS-Gα complexes from Gα<sub>i</sub> subfamily members, these residues classified into two major groups: <scene name='70/701447/Rgs4-ga-sandc-residues/4'>Significant and Conserved residues</scene> shown as red spheres that are located mainly in the center of the RGS domain–Gα interface and have a primary role in accelerating Gα GTPase by stabilizing Gα in an optimal conformation for GTP hydrolysis. Whereas the <scene name='70/701447/Rgs4-ga_modulatory_residues/2'>putative Modulatory residues</scene> shown as purple spheres are located mostly at the periphery of the interface where they contribute to Gα subunit's recognition.<ref name="Mickey2011" /> | Many RGS protein residues located in the vicinity of the <scene name='70/701447/Rgs4-ga_interface/4'>RGS domain–Gα interface</scene> (RGS protein shown as wheat cartoon and Gα<sub>i1</sub> subunit shown as white surface) contribute to RGS-G proteins interaction. Based on energy calculation and experimental validation of RGS-Gα complexes from Gα<sub>i</sub> subfamily members, these residues classified into two major groups: <scene name='70/701447/Rgs4-ga-sandc-residues/4'>Significant and Conserved residues</scene> shown as red spheres that are located mainly in the center of the RGS domain–Gα interface and have a primary role in accelerating Gα GTPase by stabilizing Gα in an optimal conformation for GTP hydrolysis. Whereas the <scene name='70/701447/Rgs4-ga_modulatory_residues/2'>putative Modulatory residues</scene> shown as purple spheres are located mostly at the periphery of the interface where they contribute to Gα subunit's recognition.<ref name="Mickey2011" /> | ||
| - | On the other side, Gα subunits participate in a range of interactions with a variety of other proteins. Therefore, they have interfaces that interact selectively with receptors, effector subfamilies and RGS proteins. However, <scene name='70/701447/Gi-rgs4_interface/4'>Gα residues</scene> from Gα<sub>i</sub> subfamily members that interact specifically with RGS proteins are highly conserved (red spheres). These Gα Residues located on Gα switch regions interact with Significant & Conserved RGS residues because of the pivotal role of the switch regions in GTP hydrolysis that is catalyzed by RGS proteins. On the other hand, Gα residues located in switch regions II and III and multiple residues in the Gα all-helical domain interact with Modulatory RGS residues.<ref name="Mickey2011" /> For example, one important conserved RGS4 residue that projects into the active site of Gα<sub>i</sub> is <scene name='70/701447/Gi-rgs4-asn128/ | + | On the other side, Gα subunits participate in a range of interactions with a variety of other proteins. Therefore, they have interfaces that interact selectively with receptors, effector subfamilies and RGS proteins. However, <scene name='70/701447/Gi-rgs4_interface/4'>Gα residues</scene> from Gα<sub>i</sub> subfamily members that interact specifically with RGS proteins are highly conserved (red spheres). These Gα Residues located on Gα switch regions interact with Significant & Conserved RGS residues because of the pivotal role of the switch regions in GTP hydrolysis that is catalyzed by RGS proteins. On the other hand, Gα residues located in switch regions II and III and multiple residues in the Gα all-helical domain interact with Modulatory RGS residues.<ref name="Mickey2011" /> For example, one important conserved RGS4 residue that projects into the active site of Gα<sub>i</sub> is <scene name='70/701447/Gi-rgs4-asn128/2'>Asn-128</scene> shown as blue sticks, which contacts the side chains of three Gα residues: Lys-180, Gln-204, and Glu-207 shown as green, magenta, and red sticks respectively.<ref name="Tesmer97" /> A unique modulatory RGS4 residue that interacts with Gα<sub>i</sub> is <scene name='70/701447/Gi-rgs4_arg166/6'>Arg-166</scene> shown as blue sticks which forms salt bridge with the positively charged side chain of Glu-116 shown as magenta sticks located in the helical domain of Gα subunit. |
| + | |||
| + | ==3D structures of regulator of G-protein signaling== | ||
| + | [[Regulator of G-protein signaling 3D structures]] | ||
| + | |||
</StructureSection> | </StructureSection> | ||
| + | |||
== References == | == References == | ||
<references/> | <references/> | ||
| + | [[Category:Topic Page]] | ||
Current revision
Regulator of G protein signaling (RGS) interactions with G proteins – RGS4-Gαi as a model structure.
| |||||||||||
References
- ↑ Milligan G, Kostenis E. Heterotrimeric G-proteins: a short history. Br J Pharmacol. 2006 Jan;147 Suppl 1:S46-55. PMID:16402120 doi:http://dx.doi.org/10.1038/sj.bjp.0706405
- ↑ 2.0 2.1 2.2 Kosloff M, Travis AM, Bosch DE, Siderovski DP, Arshavsky VY. Integrating energy calculations with functional assays to decipher the specificity of G protein-RGS protein interactions. Nat Struct Mol Biol. 2011 Jun 19;18(7):846-53. doi: 10.1038/nsmb.2068. PMID:21685921 doi:http://dx.doi.org/10.1038/nsmb.2068
- ↑ Gibbons DL, Abeler-Dorner L, Raine T, Hwang IY, Jandke A, Wencker M, Deban L, Rudd CE, Irving PM, Kehrl JH, Hayday AC. Cutting Edge: Regulator of G protein signaling-1 selectively regulates gut T cell trafficking and colitic potential. J Immunol. 2011 Sep 1;187(5):2067-71. doi: 10.4049/jimmunol.1100833. Epub 2011, Jul 27. PMID:21795595 doi:http://dx.doi.org/10.4049/jimmunol.1100833
- ↑ Jiang H, Xie Y, Abel PW, Wolff DW, Toews ML, Panettieri RA Jr, Casale TB, Tu Y. Regulator of G-protein signaling 2 repression exacerbates airway hyper-responsiveness and remodeling in asthma. Am J Respir Cell Mol Biol. 2015 Jul;53(1):42-9. doi: 10.1165/rcmb.2014-0319OC. PMID:25368964 doi:http://dx.doi.org/10.1165/rcmb.2014-0319OC
- ↑ Tatenhorst L, Senner V, Puttmann S, Paulus W. Regulators of G-protein signaling 3 and 4 (RGS3, RGS4) are associated with glioma cell motility. J Neuropathol Exp Neurol. 2004 Mar;63(3):210-22. PMID:15055445
- ↑ Chen X, Dunham C, Kendler S, Wang X, O'Neill FA, Walsh D, Kendler KS. Regulator of G-protein signaling 4 (RGS4) gene is associated with schizophrenia in Irish high density families. Am J Med Genet B Neuropsychiatr Genet. 2004 Aug 15;129B(1):23-6. PMID:15274033 doi:http://dx.doi.org/10.1002/ajmg.b.30078
- ↑ Holobotovskyy V, Manzur M, Tare M, Burchell J, Bolitho E, Viola H, Hool LC, Arnolda LF, McKitrick DJ, Ganss R. Regulator of G-protein signaling 5 controls blood pressure homeostasis and vessel wall remodeling. Circ Res. 2013 Mar 1;112(5):781-91. doi: 10.1161/CIRCRESAHA.111.300142. Epub 2013, Jan 9. PMID:23303165 doi:http://dx.doi.org/10.1161/CIRCRESAHA.111.300142
- ↑ Stewart A, Maity B, Wunsch AM, Meng F, Wu Q, Wemmie JA, Fisher RA. Regulator of G-protein signaling 6 (RGS6) promotes anxiety and depression by attenuating serotonin-mediated activation of the 5-HT(1A) receptor-adenylyl cyclase axis. FASEB J. 2014 Apr;28(4):1735-44. doi: 10.1096/fj.13-235648. Epub 2014 Jan 13. PMID:24421401 doi:http://dx.doi.org/10.1096/fj.13-235648
- ↑ Balasubramanian N, Levay K, Keren-Raifman T, Faurobert E, Slepak VZ. Phosphorylation of the regulator of G protein signaling RGS9-1 by protein kinase A is a potential mechanism of light- and Ca2+-mediated regulation of G protein function in photoreceptors. Biochemistry. 2001 Oct 23;40(42):12619-27. PMID:11601986
- ↑ Miao R, Lu Y, Xing X, Li Y, Huang Z, Zhong H, Huang Y, Chen AF, Tang X, Li H, Cai J, Yuan H. Regulator of G-Protein Signaling 10 Negatively Regulates Cardiac Remodeling by Blocking Mitogen-Activated Protein Kinase-Extracellular Signal-Regulated Protein Kinase 1/2 Signaling. Hypertension. 2016 Jan;67(1):86-98. doi: 10.1161/HYPERTENSIONAHA.115.05957. Epub , 2015 Nov 16. PMID:26573707 doi:http://dx.doi.org/10.1161/HYPERTENSIONAHA.115.05957
- ↑ Yuan X, Cao J, Liu T, Li YP, Scannapieco F, He X, Oursler MJ, Zhang X, Vacher J, Li C, Olson D, Yang S. Regulators of G protein signaling 12 promotes osteoclastogenesis in bone remodeling and pathological bone loss. Cell Death Differ. 2015 Dec;22(12):2046-57. doi: 10.1038/cdd.2015.45. Epub 2015, Apr 24. PMID:25909889 doi:http://dx.doi.org/10.1038/cdd.2015.45
- ↑ Willard FS, Willard MD, Kimple AJ, Soundararajan M, Oestreich EA, Li X, Sowa NA, Kimple RJ, Doyle DA, Der CJ, Zylka MJ, Snider WD, Siderovski DP. Regulator of G-protein signaling 14 (RGS14) is a selective H-Ras effector. PLoS One. 2009;4(3):e4884. doi: 10.1371/journal.pone.0004884. Epub 2009 Mar 25. PMID:19319189 doi:http://dx.doi.org/10.1371/journal.pone.0004884
- ↑ Pashkov V, Huang J, Parameswara VK, Kedzierski W, Kurrasch DM, Tall GG, Esser V, Gerard RD, Uyeda K, Towle HC, Wilkie TM. Regulator of G protein signaling (RGS16) inhibits hepatic fatty acid oxidation in a carbohydrate response element-binding protein (ChREBP)-dependent manner. J Biol Chem. 2011 Apr 29;286(17):15116-25. doi: 10.1074/jbc.M110.216234. Epub, 2011 Feb 27. PMID:21357625 doi:http://dx.doi.org/10.1074/jbc.M110.216234
- ↑ Hayes MP, Roman DL. Regulator of G Protein Signaling 17 as a Negative Modulator of GPCR Signaling in Multiple Human Cancers. AAPS J. 2016 May;18(3):550-9. doi: 10.1208/s12248-016-9894-1. Epub 2016 Feb 29. PMID:26928451 doi:http://dx.doi.org/10.1208/s12248-016-9894-1
- ↑ Alshbool FZ, Karim ZA, Vemana HP, Conlon C, Lin OA, Khasawneh FT. The regulator of G-protein signaling 18 regulates platelet aggregation, hemostasis and thrombosis. Biochem Biophys Res Commun. 2015 Jul 10;462(4):378-82. doi:, 10.1016/j.bbrc.2015.04.143. Epub 2015 May 9. PMID:25969426 doi:http://dx.doi.org/10.1016/j.bbrc.2015.04.143
- ↑ 16.0 16.1 Tesmer JJ, Berman DM, Gilman AG, Sprang SR. Structure of RGS4 bound to AlF4--activated G(i alpha1): stabilization of the transition state for GTP hydrolysis. Cell. 1997 Apr 18;89(2):251-61. PMID:9108480
Proteopedia Page Contributors and Editors (what is this?)
Ali Asli, Denise Salem, Michal Harel, Joel L. Sussman, Jaime Prilusky