Inosine monophosphate dehydrogenase
From Proteopedia
(Difference between revisions)
| (9 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load='Figure_2.pdb' size=' | + | <StructureSection load='Figure_2.pdb' size='350' side='right' scene='Journal:JBSD:1/Cv/1' caption='Human IMPDH ([[1b3o]])'> |
== Function == | == Function == | ||
| - | '''Inosine monophosphate dehydrogenase''' (IMPDH) is part of the GTP biosynthesis and catalyzes the conversion of inosine monophosphate to xanthosine monophosphate while reducing NAD. IMPDH is inhibited by ribavirin and mycophenolic acid. | + | '''Inosine monophosphate dehydrogenase''' (IMPDH) is part of the GTP biosynthesis and catalyzes the conversion of inosine monophosphate to xanthosine monophosphate while reducing [[NAD]]. IMPDH is inhibited by ribavirin and mycophenolic acid<ref>PMID:8681386</ref>. |
== Disease == | == Disease == | ||
| - | + | IMPDH level of activity is suggested to determine whether acute leukemia cells proliferate or differentiate<ref>PMID:2885446</ref>. | |
| - | + | ||
== Structural highlights == | == Structural highlights == | ||
| - | === An Insight to the Dynamics of Conserved Water Mediated Salt Bridge Interaction and Inter-Domain Recognition in hIMPDH Isoforms | + | === An Insight to the Dynamics of Conserved Water Mediated Salt Bridge Interaction and Inter-Domain Recognition in hIMPDH Isoforms <ref>DOI 10.1080/07391102.2012.712458</ref>=== |
| - | + | ||
| - | + | ||
| - | + | ||
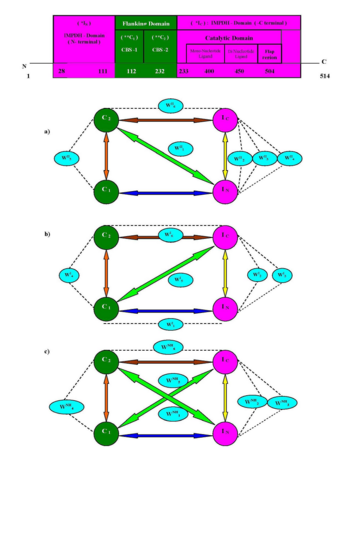
Dynamic personality of protein is inherent in Nature and necessary for performing the chemical reaction in living cells. The functions of proteins / enzymes and their physical properties are also governed by their dynamic characters which requires addition of a fourth dimension, time and atomic resolution. Detail investigation of dynamic and X-ray structures of protein or their complexes with different molecules can unfold the structure-function relationship of protein. Presumably, the dynamic propensity of protein is not only determine the chemical transformations/ reaction mechanism of biosynthetic processes but it also vividly illustrated the insight of topological feature of inhibitor. It is interesting in enzymology that Nature also installs some similar proteins which are almost have identical function, catalytic machinery, 3D-structures and are known as isoform. The salt bridge interaction (acidic and basic residues) as well as their conjugation through amphoteric water molecules (Acid---Water---Base) may be indispensable component of protein recognition, domain assemble and structure stabilization. The differential inter domain recognition through conserved water mediated salt bridge interaction in type -I and II isoforms (of human IMPDH) may open a new avenue towards their inhibitor development. | Dynamic personality of protein is inherent in Nature and necessary for performing the chemical reaction in living cells. The functions of proteins / enzymes and their physical properties are also governed by their dynamic characters which requires addition of a fourth dimension, time and atomic resolution. Detail investigation of dynamic and X-ray structures of protein or their complexes with different molecules can unfold the structure-function relationship of protein. Presumably, the dynamic propensity of protein is not only determine the chemical transformations/ reaction mechanism of biosynthetic processes but it also vividly illustrated the insight of topological feature of inhibitor. It is interesting in enzymology that Nature also installs some similar proteins which are almost have identical function, catalytic machinery, 3D-structures and are known as isoform. The salt bridge interaction (acidic and basic residues) as well as their conjugation through amphoteric water molecules (Acid---Water---Base) may be indispensable component of protein recognition, domain assemble and structure stabilization. The differential inter domain recognition through conserved water mediated salt bridge interaction in type -I and II isoforms (of human IMPDH) may open a new avenue towards their inhibitor development. | ||
| Line 38: | Line 35: | ||
Stereospecific interaction or recognition of IN to C2 domain through <scene name='Journal:JBSD:1/Cv/12'>conserved water mediated salt bridge (K109 (NZ) --- D215 / D216 and K109 (NZ) --- WII1 --- D215 / D216)</scene> are observed to be unique in hIMPDH–II, which is not observed in type I isoform (1JCN). The geometrical and electronic consequences of <scene name='Journal:JBSD:1/Cv/14'>conserved water molecular interaction as shown in Figure 5 (K109 to acidic D215 / D216 and E217)</scene> and their stereo chemical features (specially in CBS --- IN inter-domain recognition site) may be used to design the actual topology of inhibitor for hIMPDH-II isoform using water mimic inhibitor design protocol. Possibly, heterocyclic ligand with flexible structure containing two or three basic and hydrophilic groups with suitable spacer length may be implemented to design the isoform selective inhibitor for CML cancer. | Stereospecific interaction or recognition of IN to C2 domain through <scene name='Journal:JBSD:1/Cv/12'>conserved water mediated salt bridge (K109 (NZ) --- D215 / D216 and K109 (NZ) --- WII1 --- D215 / D216)</scene> are observed to be unique in hIMPDH–II, which is not observed in type I isoform (1JCN). The geometrical and electronic consequences of <scene name='Journal:JBSD:1/Cv/14'>conserved water molecular interaction as shown in Figure 5 (K109 to acidic D215 / D216 and E217)</scene> and their stereo chemical features (specially in CBS --- IN inter-domain recognition site) may be used to design the actual topology of inhibitor for hIMPDH-II isoform using water mimic inhibitor design protocol. Possibly, heterocyclic ligand with flexible structure containing two or three basic and hydrophilic groups with suitable spacer length may be implemented to design the isoform selective inhibitor for CML cancer. | ||
| - | </StructureSection> | ||
==3D structures of inosine monophosphate dehydrogenase== | ==3D structures of inosine monophosphate dehydrogenase== | ||
| + | [[Inosine monophosphate dehydrogenase 3D structures]] | ||
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | }} | ||
== References == | == References == | ||
<references/> | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
References
- ↑ Sintchak MD, Fleming MA, Futer O, Raybuck SA, Chambers SP, Caron PR, Murcko MA, Wilson KP. Structure and mechanism of inosine monophosphate dehydrogenase in complex with the immunosuppressant mycophenolic acid. Cell. 1996 Jun 14;85(6):921-30. PMID:8681386
- ↑ Price GM, Hoffbrand AV, Taheri MR, Evans JP. Inosine monophosphate dehydrogenase activity in acute leukaemia. Leuk Res. 1987;11(6):525-8. PMID:2885446
- ↑ Bairagya HR, Mukhopadhyay BP. An insight to the dynamics of conserved water-mediated salt bridge interaction and interdomain recognition in hIMPDH isoforms. J Biomol Struct Dyn. 2012 Aug 28. PMID:22928911 doi:10.1080/07391102.2012.712458