This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
NADP-dependent malic enzyme
From Proteopedia
(Difference between revisions)
(New page: ==Your Heading Here (maybe something like 'Structure')== <StructureSection load='1stp' size='340' side='right' caption='Caption for this structure' scene=''> This is a default text for you...) |
|||
| (78 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
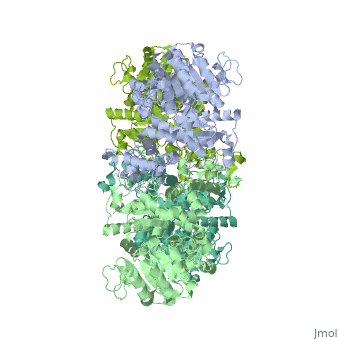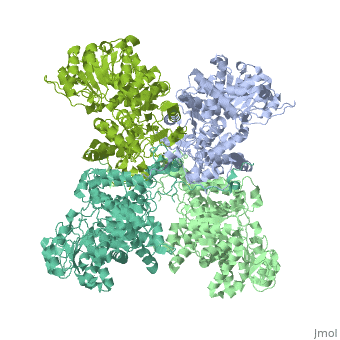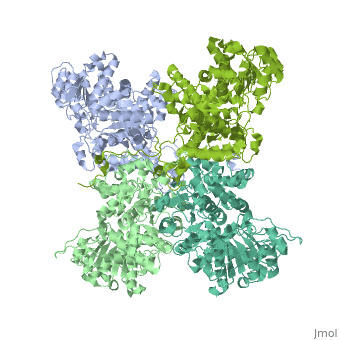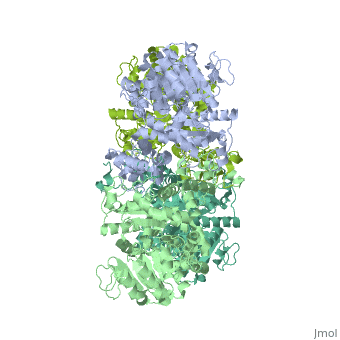
| - | + | <StructureSection load='3wja' size='340' side='right' caption='The best structure for NADP-dependent malic enzyme shown: [[3wja]]' scene=''> | |
| - | <StructureSection load=' | + | Best example is PDB entry [[3wja]] and is shown in the viewer.<br> |
| - | + | ==Catalytic Activity == | |
| - | + | Oxaloacetate = pyruvate + CO(2).Data source: Uniprot [http://www.uniprot.org/uniprot/P48163 P48163]<br> | |
| + | Or the decarboxylation of malate to pyruvate<ref>PMID:8187880</ref>. | ||
| - | == | + | == Biological process == |
| + | Is involved in the following biological processes:<br> | ||
| + | malate metabolic process<br> | ||
| + | oxidation-reduction process<br> | ||
| + | response to carbohydrate<br> | ||
| + | response to hormone<br> | ||
| + | carbohydrate metabolic process<br> | ||
| + | protein tetramerization<br> | ||
| + | regulation of NADP metabolic process<br> | ||
| + | NADP biosynthetic process<br> | ||
| + | small molecule metabolic process<br> | ||
| + | cellular lipid metabolic process<br> | ||
| + | == In structures == | ||
| + | NADP-dependent malic enzyme is found in 3 PDB entries<br> | ||
| + | *<b>Eukaryota</b> (3 PDB entries): <br> | ||
| + | **<b>Homo sapiens</b> (2 PDB entries): <br> | ||
| + | ***[[3wja]] <div class="pdb-prints 3wja"></div> | ||
| + | ***Title: The crystal structure of human cytosolic NADP(+)-dependent malic enzyme in apo form | ||
| + | ***2.548 A resolution | ||
| + | ***[http://www.ebi.ac.uk/pdbe/entry/search/index?organism_scientific_name:%22Homo%20sapiens%22&all_molecule_names:%22NADP-dependent%20malic%20enzyme%22&!chimera:y Search the PDB for NADP-dependent malic enzyme from Homo sapiens] | ||
| + | **<b>Columba livia</b> (1 PDB entries): <br> | ||
| + | ***[[1gq2]] <div class="pdb-prints 1gq2"></div> | ||
| + | ***Title: MALIC ENZYME FROM PIGEON LIVER | ||
| + | ***2.5 A resolution | ||
| + | == Alternative names for NADP-dependent malic enzyme == | ||
| + | Molecule '''NADP-dependent malic enzyme''', also known as '''NADP-ME''', '''NADP-dependent malic enzyme''' and '''Malic enzyme 1'''. | ||
| - | == | + | == 3D Structures of NADP-dependent malic enzyme == |
| - | + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | |
| + | |||
| - | + | [[3wja]], [[2aw5]] – hME - human<br /> | |
| + | [[7x11]], [[7x12]] – hME1 + NADP<br /> | ||
| + | [[8e76]], [[8eyn]] – hME3<br /> | ||
| + | [[8e78]], [[8e8o]], [[8eyo]] – hME3 + NADP<br /> | ||
| + | [[1gq2]] – ME + NADP - pigeon<br /> | ||
| + | [[5ou5]] – ME - maize<br /> | ||
| - | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
| - | |||
| - | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
| + | [[Category:Topic Page]][[Category:PDBe]] | ||
Current revision
| |||||||||||