User:Jeremiah C Hagler/Protein 1
From Proteopedia
< User:Jeremiah C Hagler(Difference between revisions)
(→Introduction to Computer-Aided Protein Visualization Lab) |
|||
| (7 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
== Introduction to Computer-Aided Protein Visualization Lab == | == Introduction to Computer-Aided Protein Visualization Lab == | ||
<StructureSection load='1pgb' size='340' side='right' caption='This simple protein, B1 Immunoglobulin-binding domain of Streptococcal protein G, shows secondary structures nicely. The alpha helix is red, beta sheet in yellow.' scene='71/713432/Protein_secondary_structure/1'> | <StructureSection load='1pgb' size='340' side='right' caption='This simple protein, B1 Immunoglobulin-binding domain of Streptococcal protein G, shows secondary structures nicely. The alpha helix is red, beta sheet in yellow.' scene='71/713432/Protein_secondary_structure/1'> | ||
| + | |||
== Computer-Aided Protein Visualization Lab == | == Computer-Aided Protein Visualization Lab == | ||
Knowing the three-dimensional structure of a protein can be a very powerful tool for biologists. Much can be learned about enzyme function, interaction of molecules in your immune system, the appearance of the surface of viruses, and the interaction of ligands and receptors. | Knowing the three-dimensional structure of a protein can be a very powerful tool for biologists. Much can be learned about enzyme function, interaction of molecules in your immune system, the appearance of the surface of viruses, and the interaction of ligands and receptors. | ||
| Line 6: | Line 7: | ||
== First some background: (make sure that you understand the underlined words)== | == First some background: (make sure that you understand the underlined words)== | ||
| - | Proteins are synthesized on ribosomes by linking together many amino acids into a long chain. If you could observe a protein as it is made, it would look like a string of pearls (amino acids) feeding out the end of the ribosome as it floats in the cytoplasm of the cell [https://www.dnalc.org/view/15501-Translation-RNA-to-protein-3D-animation-with-basic-narration.html Video of Translation (DNALC)]. This structure is called the primary structure (or 1° structure) and refers to the sequence of amino acids of the protein. After protein synthesis has started, two choices are possible: | + | Proteins are synthesized on ribosomes by linking together many amino acids into a long chain. If you could observe a protein as it is made, it would look like a string of pearls (amino acids) feeding out the end of the ribosome as it floats in the cytoplasm of the cell ([https://www.dnalc.org/view/15501-Translation-RNA-to-protein-3D-animation-with-basic-narration.html Video of Translation (DNALC)]). This structure is called the primary structure (or 1° structure) and refers to the sequence of amino acids of the protein. After protein synthesis has started, two choices are possible: |
(1) if the protein is destined to be secreted or to reside in an organelle of the secretory pathway, the first twenty or so amino acids will comprise a signal sequence. These act to direct the ribosome to the endoplasmic reticulum (ER) where the protein will be fed through a channel in the membrane into the interior of the ER. Once inside the ER, the protein will fold and receive sugar modifications called glycosylations. | (1) if the protein is destined to be secreted or to reside in an organelle of the secretory pathway, the first twenty or so amino acids will comprise a signal sequence. These act to direct the ribosome to the endoplasmic reticulum (ER) where the protein will be fed through a channel in the membrane into the interior of the ER. Once inside the ER, the protein will fold and receive sugar modifications called glycosylations. | ||
| Line 22: | Line 23: | ||
<br> | <br> | ||
The second step of protein folding results in the tertiary structure (or 3° structure). Tertiary structure gives the protein an overall three-dimensional structure. The tertiary structure of a protein is determined by a combination of factors including hydrogen bonds, ionic bonds (between positively and negatively charged amino acids), covalent disulfide bonds (between cysteine residues), and Van der Waals interactions. Tertiary structure can also be affected by repulsive forces between similarly charged amino acids, as well as hydrophobic and hydrophilic interactions with a solvent (commonly water). At a distance many proteins form what look to be large globs at this point, and it is only upon more careful and close up inspection that one can see the true uniqueness of the shape. | The second step of protein folding results in the tertiary structure (or 3° structure). Tertiary structure gives the protein an overall three-dimensional structure. The tertiary structure of a protein is determined by a combination of factors including hydrogen bonds, ionic bonds (between positively and negatively charged amino acids), covalent disulfide bonds (between cysteine residues), and Van der Waals interactions. Tertiary structure can also be affected by repulsive forces between similarly charged amino acids, as well as hydrophobic and hydrophilic interactions with a solvent (commonly water). At a distance many proteins form what look to be large globs at this point, and it is only upon more careful and close up inspection that one can see the true uniqueness of the shape. | ||
| - | + | <br> | |
| - | + | [[Image:Intramolecular forces in tertiary structures.png]] | |
Figure 3: The various intramolecular interactions that help determine teriary structure | Figure 3: The various intramolecular interactions that help determine teriary structure | ||
| Line 35: | Line 36: | ||
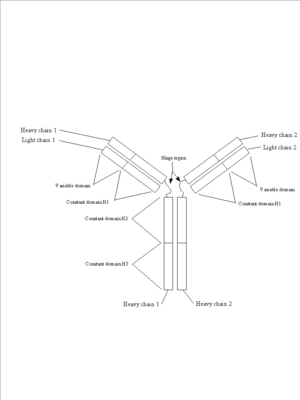
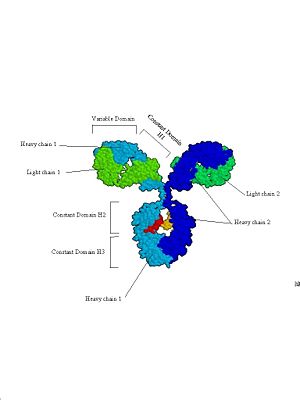
Figure 4: Immunoglobulins. Three views of the immunoglobulin complex IgG. | Figure 4: Immunoglobulins. Three views of the immunoglobulin complex IgG. | ||
<br> | <br> | ||
| - | a. [[Image:Antibody schematic.png]] | + | a. [[Image:Antibody schematic.png|300px|]] |
<br> | <br> | ||
This is a schematic of an immunoglobulin complex showing the two light chains, the two heavy chains and the variable and constant domains. | This is a schematic of an immunoglobulin complex showing the two light chains, the two heavy chains and the variable and constant domains. | ||
<br> | <br> | ||
| - | b. [[Image:Antibody spacefilling diagram.jpg]] | + | b. [[Image:Antibody spacefilling diagram.jpg|300px|]] |
<br> | <br> | ||
This is a space filling model based on an X-ray crystal structure. This clearly shows the 4 subunits that make up antibodies, and how they are divided into functional (and structural) domains. | This is a space filling model based on an X-ray crystal structure. This clearly shows the 4 subunits that make up antibodies, and how they are divided into functional (and structural) domains. | ||
| Line 59: | Line 60: | ||
This now brings us back to where we started at the top of page 1 - once we know the structure, how can we look at it? There are many computer programs in existence to visualize proteins in three-dimensions. For the drug design studies, powerful computers and programs are necessary to analyze the energetics of drug fit. (Simulating structures and their interactions is a powerful “weeding out” tool in deciding which drugs to test in laboratory studies, making the development of new drugs more efficient and less costly.) | This now brings us back to where we started at the top of page 1 - once we know the structure, how can we look at it? There are many computer programs in existence to visualize proteins in three-dimensions. For the drug design studies, powerful computers and programs are necessary to analyze the energetics of drug fit. (Simulating structures and their interactions is a powerful “weeding out” tool in deciding which drugs to test in laboratory studies, making the development of new drugs more efficient and less costly.) | ||
| - | For this | + | For this lab you will be working with a very commonly used computer modeling program known as Jsmol to look at "pdb" files of proteins with a known structure (using the methods outlined above). Jsmol is an open-source JavaScript viewer for chemical structures in 3D: http://wiki.jmol.org/index.php/JSmol. We are using the website proteopedia.org as a wiki host site to house and access these exercises. |
| + | |||
== Preliminary Questions == | == Preliminary Questions == | ||
| Line 76: | Line 78: | ||
Make sure you answer all questions in the lab. Questions marked by an * are more complicated and may require outside reading and thinking time to answer completely. | Make sure you answer all questions in the lab. Questions marked by an * are more complicated and may require outside reading and thinking time to answer completely. | ||
<br> | <br> | ||
| - | == == | + | == Using Jmol through proteopedia.org == |
| + | A VERY BRIEF GUIDE TO USING Jmol | ||
| + | |||
| + | Logon to your computer, then navigate to the following website: | ||
| + | |||
| + | All of the structures hosted these sites initially are viewable in the small window on the right side of the page. However, if you click on "popup" in the lower left of that window, a larger window containing the structure will become visible. This window can be resized to any size by dragging the window edges to desirable dimensions. | ||
| + | <br> | ||
| + | TURNING THE PROTEIN: Once you have the molecule displayed on the black screen, try rotating the molecule by placing the pointer on the screen, holding down the mouse button and dragging the mouse. Move the mouse slowly so that you can see the molecule turn. If you place the pointer in the center of the field and drag left/right, the protein will rotate around the y-axis. If you place the pointer in the center of the field and drag up/down, the protein will rotate around the x-axis. If you place the pointer at the top of the field and drag left/right, the protein will rotate around the z-axis. | ||
| + | |||
| + | TO VIEW SELECTED GROUPS (carbon backbones or side chains of amino acids): Make sure the protein is not spinning (click on "toggle spin" in the bottom left corner of the image window to stop spinning). Then, simply move your cursor to any portion of the molecule you want to identify. After a second or two, a small window should popup containing a chain of information--a three letter code (amino acid) followed by a number (the position of the amino acid in the protein primary structure), a chain designation (indicating which protein chain/subunit you are pointing at) and then the element you are pointing at (C,H,N,O,P,S,Zn,Mg,Ca,etc). | ||
| + | |||
| + | == Here are the structures you will want to examine == | ||
| + | 1.[http://proteopedia.org/wiki/index.php/User:Jeremiah_C_Hagler/Sandbox_2 Protein 1: MHC Class I] | ||
| + | <br> | ||
| + | 2.[http://proteopedia.org/wiki/index.php/User:Jeremiah_C_Hagler/Sandbox_3 Protein 3: HIV Reverse Transcriptase] | ||
| + | <br> | ||
| + | 3.[http://proteopedia.org/wiki/index.php/User:Jeremiah_C_Hagler/Sandbox_1 Protein 2: Alkaline Phosphatase] | ||
| + | <br> | ||
| + | |||
<references/> | <references/> | ||
Current revision
Introduction to Computer-Aided Protein Visualization Lab
| |||||||||||