Timothy Locksmith sandbox Heat Sock Factor
From Proteopedia
(Difference between revisions)
| (32 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
<StructureSection load='3hts' size='340' side='right' caption='Caption for this structure' scene=''> | <StructureSection load='3hts' size='340' side='right' caption='Caption for this structure' scene=''> | ||
| - | This is a default text for your page '''Timothy Locksmith sandbox Heat Shock Factor'''. Click above on '''edit this page''' to modify. | ||
| - | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:10331875</ref> to the rescue. | ||
| - | |||
== Introduction == | == Introduction == | ||
| - | Heat Shock Factor (HSF) is primarily used for the transcription of Heat shock proteins, which are used | + | Heat Shock Factor (HSF) is primarily used for the transcription of Heat shock proteins, which are used for the survival of a cell under high temperature stress situations.<ref>PMID: 24800749</ref> Its structure is highly conserved through species from flies to humans, and even all the way to yeast, which suggests that its existence is very important for the survival of organisms. Studies delving into the uses of HSF have proven promising in Cancer research fields. In some instances when cancer cells had their HSF inhibited underwent cell death instead of proliferation, thus ceasing the spread of the cancer and reducing its size. It is hypothesized that HSF is an integral part of cancer cell survival. |
== Regulation == | == Regulation == | ||
| - | HSF is inhibited by heat shock proteins under normal conditions (regarding temperature). The main regulator of HSF is the temperature of the surrounding environment. When the temperature is high, Hsp’s then associate elsewhere the in the cell with damages structures, and are therefore less likely to bind to HSF. This allows HSP to | + | HSF is inhibited by heat shock proteins under normal conditions (regarding temperature). The main regulator of HSF is the temperature of the surrounding environment. When the temperature is high, Hsp’s then associate elsewhere the in the cell with damages structures, and are therefore less likely to bind to HSF. This allows HSP to move to the inside of the nucleus, and interact with it to produce more Hsp’s. Once enough Hsp’s are created, the damage has been taken care of and the temperature has returned to normal, they can once again bind to HSF, and inhibit it from interacting with the DNA. |
== Sturcture/DNA Interaction == | == Sturcture/DNA Interaction == | ||
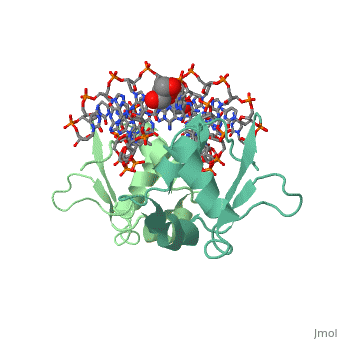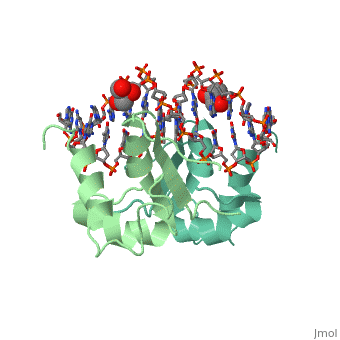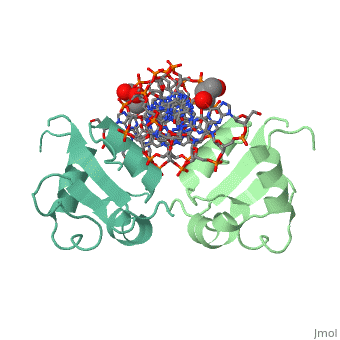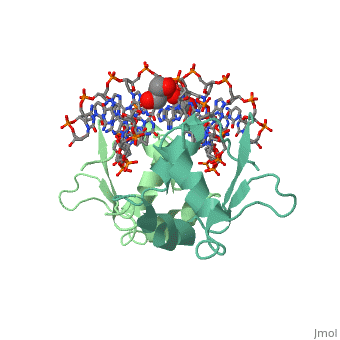
| - | HSF remains as a monomer when bound with Hsp under non-stressed conditions. Under stressed conditions (increased temperature) three individual monomers of HSF move into the cell’s nucleus, | + | HSF remains as a monomer when bound with Hsp under non-stressed conditions. Under stressed conditions (increased temperature) three individual monomers of HSF move into the cell’s nucleus, <scene name='71/714949/Dimer_hsf_with_dna_interaction/3'>trimerizes</scene> (could only isolate a dimer structure), and then bind to the Major Groove of the DNA though both hydrogen bonding, and it is believed that the residues are close enough for van der Waals interaction.<ref>PMID: 8421783</ref> Specifically an <scene name='71/714949/Arg_met_interact/2'>Arginine, and Methionine</scene> from each of the monomer bind to <scene name='71/714949/Alternating_gaa/2'>three oppositely oriented</scene> "<scene name='71/714949/Gaa_met_arg_interact/2'>nGAAn</scene>" sections of the genome.<ref>PMID:10331875</ref> These residues interact with the nucleotide bases, while nearby residues interact with the back bone to stabilize the transcription factor in place. The structure is classified as a <scene name='71/714949/Dimer_hsf_with_dna_interaction/5'>"winged" helix turn helix</scene> protein. These proteins usually consist of three alpha helices and three beta sheets which are arranged so that one helix interacts with the major groove of DNA while the "wings" interact with the minor groove for further DNA interaction or the backbone for specificity in binding. <ref>PMID:10679470</ref> However this was found to not be the case with HSF, instead the "wings" are used to form the dimer with a second monomer of HSF.[3] |
| - | + | ||
| - | + | ||
| - | </ | + | </structuresection> |
== References == | == References == | ||
<references/> | <references/> | ||
Current revision
| |||||||||||
References
- ↑ Salamanca HH, Antonyak MA, Cerione RA, Shi H, Lis JT. Inhibiting heat shock factor 1 in human cancer cells with a potent RNA aptamer. PLoS One. 2014 May 6;9(5):e96330. doi: 10.1371/journal.pone.0096330. eCollection , 2014. PMID:24800749 doi:http://dx.doi.org/10.1371/journal.pone.0096330
- ↑ Rabindran SK, Haroun RI, Clos J, Wisniewski J, Wu C. Regulation of heat shock factor trimer formation: role of a conserved leucine zipper. Science. 1993 Jan 8;259(5092):230-4. PMID:8421783
- ↑ Littlefield O, Nelson HC. A new use for the 'wing' of the 'winged' helix-turn-helix motif in the HSF-DNA cocrystal. Nat Struct Biol. 1999 May;6(5):464-70. PMID:10331875 doi:10.1038/8269
- ↑ Gajiwala KS, Burley SK. Winged helix proteins. Curr Opin Struct Biol. 2000 Feb;10(1):110-6. PMID:10679470