Tyrone Evans Hox Proteins sandbox
From Proteopedia
(Difference between revisions)
| (35 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
==Hox Proteins== | ==Hox Proteins== | ||
<StructureSection load='1puf' size='340' side='right' caption='Caption for this structure' scene=''> | <StructureSection load='1puf' size='340' side='right' caption='Caption for this structure' scene=''> | ||
| - | |||
| - | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
== Introduction == | == Introduction == | ||
| - | Hox Proteins are transcription factors, proteins that are involved in the process of | + | Hox Proteins are transcription factors, proteins that are involved in the process of transcribing DNA into RNA. These specific types of proteins are capable of binding to enhancers, a short (50-1500 bp) region of DNA, in pursuit of activating or repressing certain genes. These proteins are named this way because of the mutations that are done within them that cause homeotic transformations, changing of an organ into another<ref>"Hox Gene." Wikipedia. Wikimedia Foundation, 14 Aug. 2015. Web. 20 Oct. 2015.</ref> <ref>Ladam, Franck, and Charles G. Sagerström. “Hox Regulation of Transcription – More Complex(es).” Developmental dynamics : an official publication of the American Association of Anatomists 243.1 (2014): 10.1002/dvdy.23997. PMC. Web. 3 Nov. 2015.</ref> |
| + | . | ||
| - | == | + | == Role in Development == |
| - | + | As mentioned previously, homeotic transformations can alter DNA sequences so that specific proteins will have different functions, and certain genes will be either activated or deactivated. More specifically, if a certain hox protein is responsible for the development of legs on a fly in the middle region of its body, a homeotic transformation can cause this protein to carry out this same function in a different area. In this example, this altered protein can activate the gene for leg development to change the antennae to legs on that fly's head, as seen in multiple experiments. And biologists believe this to be true for Hox genes in flies, mice, humans, and other animals<ref>Freeman, Quillin, and Allison. Biological Science. 5th ed. Vol. 1. N.p.: Benjamin Cummings, n.d. Print. | |
| + | </ref>. | ||
| - | == Hox Protein | + | == Hox Protein Regulation == |
| - | Hox genes, which hold the instructions for how each protein should be constructed, are regulated by both gap genes and pair-rule genes. Gap genes are responsible for the development of particular sections of an organism, more specifically for embryos of some arthropods. If a mutation occurs with these genes, adjoining body sections can be loss. Pair-rule genes are similar to gap genes but instead of being involved in the development of arthropods, it participates in the segment development of insects. And both of these regulators are ultimately regulated by maternally-supplied mRNA. In a series of events, depending on the amount of said maternally-supplied mRNA either the gap or pair-rule genes are activated, which in turn activate the Hox genes, allowing them to cause a differentiation in the genes that are responsible for the construction of proteins for embryo development | + | Hox genes, which hold the instructions for how each protein should be constructed, are regulated by both gap genes and pair-rule genes. Gap genes are responsible for the development of particular sections of an organism, more specifically for embryos of some arthropods. If a mutation occurs with these genes, adjoining body sections can be loss. Pair-rule genes are similar to gap genes but instead of being involved in the development of arthropods, it participates in the segment development of insects. And both of these regulators are ultimately regulated by maternally-supplied mRNA. In a series of events, depending on the amount of said maternally-supplied mRNA either the gap or pair-rule genes are activated, which in turn activate the Hox genes, allowing them to cause a differentiation in the genes that are responsible for the construction of proteins for embryo development<ref>Ladam, Franck, and Charles G. Sagerström. “Hox Regulation of Transcription – More Complex(es).” Developmental dynamics : an official publication of the American Association of Anatomists 243.1 (2014): 10.1002/dvdy.23997. PMC. Web. 3 Nov. 2015.</ref>.. |
| - | + | ||
== Interaction with DNA == | == Interaction with DNA == | ||
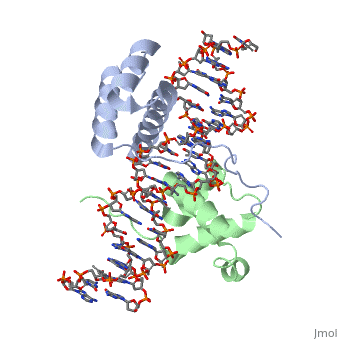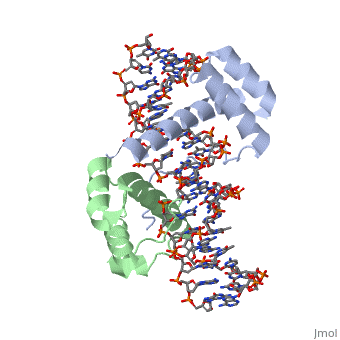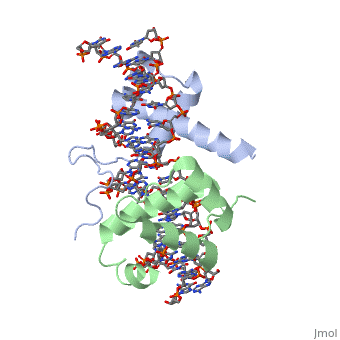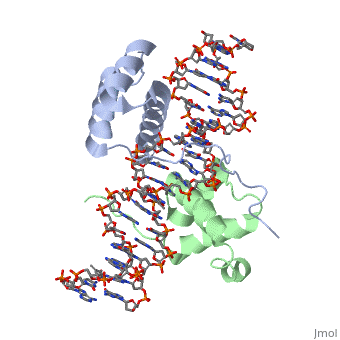
| - | A homeodomain is an essential part that all Hox proteins possess. The homeodomain can be found within the homeobox, a DNA sequence found within genes that are responsible of morphogenesis of living organisms. This sequence codes for homeodomain protein products, 60 amino-acid segments, which have specific folding patterns (helix-turn-helix) that allows them to bind with DNA through a 5’- | + | A homeodomain is an essential part that all Hox proteins possess. The homeodomain can be found within the homeobox, a DNA sequence found within genes that are responsible of morphogenesis of living organisms. This sequence codes for homeodomain protein products, 60 amino-acid segments, which have specific folding patterns (helix-turn-helix) that allows them to bind with DNA through a <scene name='71/714951/Taaat_sequence/1'>5’-TAAAT-3’</scene> core motif. This domain consists of three helical regions folded into a tight spherical structure. There are <scene name='71/714951/Both_n_and_c-terminal_helices/1'>two antiparallel N-terminal helices and one C-terminal helix</scene> (Recognition helix) within this domain. The C-terminal helix binds directly with DNA through the use of <scene name='71/714951/H_bond_between_gln_and_p/1'>Hydrogen bonds</scene> and <scene name='71/714951/Hydrophobic_interaction/3'>Hydrophobic interactions</scene> between side chains and the outer bases and thymine methyl groups within the major groove of a DNA helix. Ionic bonding can also be observed between the bases of the DNA sequence and the amino acids of the protein dimers. A more specific <scene name='71/714951/Ionic_bond/1'>Ionic interaction</scene> is the one between LYS 207 and an oxygen atom of a phosphate group. The <scene name='71/714951/Recognition_helix/1'>Recognition helix</scene> within homeodomain binds within the <scene name='71/714951/Dna_major_groove/1'>major groove</scene> of a DNA helix, while the <scene name='71/714951/Amino-terminal_tail/1'>Amino-Terminal tail</scene> binds within the <scene name='71/714951/Dna_minor_groove/1'>minor groove</scene> of a DNA helix<ref>"Hox Gene." Wikipedia. Wikimedia Foundation, 14 Aug. 2015. Web. 20 Oct. 2015.</ref>. |
| + | |||
| + | == Human Hox Genes<ref>"Hox Gene." Wikipedia. Wikimedia Foundation, 14 Aug. 2015. Web. 20 Oct. 2015.</ref> == | ||
| + | |||
| + | The cluster HOX-A corresponds with chromosome 7, regulating genes: HOX-A1, HOX-A2, HOX-A3, HOX-A4, HOX-A5, HOX-A6, HOX-A7, HOX-A9, HOXA-10, HOXA-11, HOXA-13. | ||
| + | |||
| + | The cluster HOX-B corresponds with chromosome 17, regulating genes: HOX-B1, HOX-B2, HOX-B3, HOX-B4, HOX-B5, HOX-B6, HOX-B7, HOX-B8, HOX-B9, HOX-B13. | ||
| + | |||
| + | The cluster HOX-C corresponds with chromosome 12, regulating genes:HOX-C4, HOX-C5, HOX-C6, HOX-C8, HOX-C9, HOX-C10, HOX-C11, HOX-C12, HOX-C13. | ||
| + | |||
| + | And finally, the cluster HOX-D corresponds with chromosome 2, regulating genes:HOX-D1, HOX-D3, HOX-D4, HOX-D8, HOX-D9, HOX-D10, HOX-D11, HOX-D12, HOX-D13. | ||
| + | |||
| + | == Structural highlights<ref>Berman, Helen M., John Westbrook, Zukang Feng, Gary Gilliland, T. N. Bhat, Helge Weissig, Ilya N. Shindyalov, and Philip E. Bourne. "The Protein Data Bank." RCSB. Nucl. Acids Res. (2000) 28 (1): 235-242 Web.</ref> == | ||
| + | <scene name='71/714951/Taaat_sequence/1'>5’-TAAT-3’</scene> | ||
| + | |||
| + | <scene name='71/714951/Both_n_and_c-terminal_helices/1'>Two antiparallel N-terminal helices and one C-terminal helix</scene> | ||
| + | |||
| + | <scene name='71/714951/H_bond_between_gln_and_p/1'>Hydrogen bonds</scene> | ||
| + | |||
| + | <scene name='71/714951/Hydrophobic_interaction/3'>Hydrophobic interactions</scene> | ||
| + | |||
| + | <scene name='71/714951/Ionic_bond/1'>Ionic interaction</scene> | ||
| + | |||
| + | <scene name='71/714951/Recognition_helix/1'>Recognition helix</scene> | ||
| - | = | + | <scene name='71/714951/Dna_major_groove/1'>Major Groove</scene> |
| - | + | <scene name='71/714951/Amino-terminal_tail/1'>Amino-Terminal tail</scene> | |
| - | + | <scene name='71/714951/Dna_minor_groove/1'>Minor Groove</scene> | |
| - | + | ||
| - | + | ||
| - | <scene name='71/714951/Dna_minor_groove/1'> | + | |
| - | + | ||
| - | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Current revision
Hox Proteins
| |||||||||||