We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Aconitase
From Proteopedia
(Difference between revisions)
| (17 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load='' size=' | + | <StructureSection load='' size='350' side='right' scene='Aconitase/Cv/1' caption='Bovine aconitase showing FeS4 cluster complex with sulfate (PDB code [[1amj]])'> |
==Function== | ==Function== | ||
| - | [[Aconitase]] (ACO, EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=4.2.1.3 4.2.1.3]) is an enzymatic domain that confers the ability to catalyse the equilibrium | + | [[Aconitase]] (ACO, EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=4.2.1.3 4.2.1.3]) is an enzymatic domain that confers the ability to catalyse the equilibrium |
:citrate = aconitate + H<sub>2</sub>O = L-isocitrate | :citrate = aconitate + H<sub>2</sub>O = L-isocitrate | ||
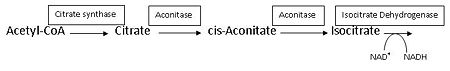
This reaction is part of the citrate (TCA-, Krebs-)cycle. | This reaction is part of the citrate (TCA-, Krebs-)cycle. | ||
In most organisms, there is a cytosolic enzyme with an ACO domain (cAc), and in eukaryotes, a second copy of it was introduced with mitochondria (mAc). Plants developed even more copies in mitochondria. | In most organisms, there is a cytosolic enzyme with an ACO domain (cAc), and in eukaryotes, a second copy of it was introduced with mitochondria (mAc). Plants developed even more copies in mitochondria. | ||
| - | Aconitase contains a Fe4S4 cluster which converts to Fe3S4 when the enzyme is inactive. In humans, two types of ACO are expressed: the soluble '''ACO1''' and the mitochondrial '''ACO2'''. | + | Aconitase contains a Fe4S4 cluster which converts to Fe3S4 when the enzyme is inactive. In humans, two types of ACO are expressed: the soluble '''ACO1''' and the mitochondrial '''ACO2'''. Two types of '''ACO X''' were characterized as '''mevalonate 5-phosphate dehydratase''' and '''cis-3-hydroxy-L-proline dehydrates'''. |
| - | Aconitase from pig (PDB [[7acn]]) is a single polypeptide (M<sub>r</sub> 83kD) that catalyzes the reversible isomerization of citrate and isocitrate.<ref name="Zheng">PMID 1313811</ref> It is the second enzyme in the Citric acid cycle, which is a series of enzyme-catalysed chemical reactions that is crucial to aerobic cellular respiration and the production of ATP. | + | Aconitase from pig (PDB [[7acn]]) is a single polypeptide (M<sub>r</sub> 83kD) that catalyzes the reversible isomerization of citrate and isocitrate.<ref name="Zheng">PMID 1313811</ref> It is the second enzyme in the Citric acid cycle, which is a series of enzyme-catalysed chemical reactions that is crucial to aerobic cellular respiration and the production of ATP. See also:<br /> |
| + | |||
| + | *[[Citric Acid Cycle]] | ||
| + | *[[Krebs cycle step 2]] | ||
| + | *[[Glyoxylate cycle]] | ||
==Structure== | ==Structure== | ||
| Line 17: | Line 21: | ||
== Catalytic mechanism of mitochondrial ACO == | == Catalytic mechanism of mitochondrial ACO == | ||
| - | Both mAc and cAc are quite similar in their ACO function. Studies, however, concentrated on <scene name='Aconitase/7acn-sf4/1'>the mitochondrial ACO</scene>. ACO is an excellent system for understanding the role of iron-sulfur-clusters in catalysis. The <scene name='Aconitase/7acn-sf4/2'>(4Fe-4S) cofactor is held in place</scene> by three sulfur atoms belonging to the cysteins-385, -448, and -451 <scene name=' | + | Both mAc and cAc are quite similar in their ACO function. Studies, however, concentrated on <scene name='Aconitase/7acn-sf4/1'>the mitochondrial ACO</scene>. ACO is an excellent system for understanding the role of iron-sulfur-clusters in catalysis. The <scene name='Aconitase/7acn-sf4/2'>(4Fe-4S) cofactor is held in place</scene> by three sulfur atoms belonging to the cysteins-385, -448, and -451 <scene name='33/338089/7acn-morph/5'>which are bound to three of the four</scene> cluster iron atoms. On activation of the enzyme, <scene name='33/338089/7acn-morph/8'>a fourth iron atom is included in the cluster</scene> together with a water molecule.This Fe4 is free to bind one, two, or three partners, in this reaction always oxygen atoms belonging to other molecules.<ref>PMID:8151704</ref> |
<!--It is clear that, in order to synthesize L-isocitrate, stereoselective catalysis must occur.--> | <!--It is clear that, in order to synthesize L-isocitrate, stereoselective catalysis must occur.--> | ||
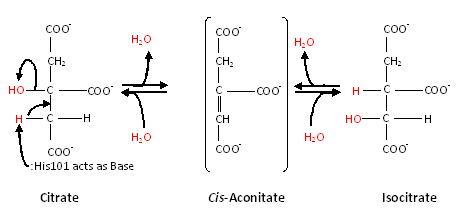
Substrate-free aconitase contains a [4Fe-4S]<sup>2+</sup> cluster with hydroxyl bound to one of the Fe. Upon binding of substrate the bound hydroxyl is protonated. A hydrogen bond from <scene name='Anthony_Noles_Sandbox/His101/3'>His101</scene> to the isocitrate hydroxyl is donated to form water. Alternatively, the proton could be donated by <scene name='Anthony_Noles_Sandbox/His167/3'>His167</scene> as this histidine is hydrogen bonded to a H<sub>2</sub>O molecule. His167 is also hydrogen bonded to the bound H<sub>2</sub>O in the [4Fe-4S] cluster. Both <scene name='Anthony_Noles_Sandbox/His_101_and_167/4'>His101 and His167</scene> are paired with carboxylates (<scene name='Anthony_Noles_Sandbox/Asp100_and_glu262/3'>Asp100 and Glu262</scene>, respectively) and are likely to be protonated. The conformational change associated with substrate binding reorients the cluster. <ref name="Beinert" /> The residue which removes a proton from citrate or isocitrate is <scene name='Anthony_Noles_Sandbox/Ser642/4'>Ser642</scene>. <ref name="Beinert" /> This causes the cis-Aconitate intermediate (seen below), which consists of a double bond, which is a direct result of the deprotonation. Then, there is a rehydration of the double bond of cis-aconitate to form isocitrate (if the original substrate was citrate). To better understand this, consider this process as stages, seen below. | Substrate-free aconitase contains a [4Fe-4S]<sup>2+</sup> cluster with hydroxyl bound to one of the Fe. Upon binding of substrate the bound hydroxyl is protonated. A hydrogen bond from <scene name='Anthony_Noles_Sandbox/His101/3'>His101</scene> to the isocitrate hydroxyl is donated to form water. Alternatively, the proton could be donated by <scene name='Anthony_Noles_Sandbox/His167/3'>His167</scene> as this histidine is hydrogen bonded to a H<sub>2</sub>O molecule. His167 is also hydrogen bonded to the bound H<sub>2</sub>O in the [4Fe-4S] cluster. Both <scene name='Anthony_Noles_Sandbox/His_101_and_167/4'>His101 and His167</scene> are paired with carboxylates (<scene name='Anthony_Noles_Sandbox/Asp100_and_glu262/3'>Asp100 and Glu262</scene>, respectively) and are likely to be protonated. The conformational change associated with substrate binding reorients the cluster. <ref name="Beinert" /> The residue which removes a proton from citrate or isocitrate is <scene name='Anthony_Noles_Sandbox/Ser642/4'>Ser642</scene>. <ref name="Beinert" /> This causes the cis-Aconitate intermediate (seen below), which consists of a double bond, which is a direct result of the deprotonation. Then, there is a rehydration of the double bond of cis-aconitate to form isocitrate (if the original substrate was citrate). To better understand this, consider this process as stages, seen below. | ||
| Line 40: | Line 44: | ||
{{Clear}} | {{Clear}} | ||
== Cytosolic aconitase and its other function == | == Cytosolic aconitase and its other function == | ||
| - | A specialty of cAc is that in mammals it has developed a <scene name=' | + | A specialty of cAc is that in mammals it has developed a <scene name='33/338089/Cv/2'>second function</scene> as inhibitor of <scene name='33/338089/Cv/3'>those mRNA</scene> that carry an <scene name='33/338089/Cv/4'>iron-responsive element (IRE)</scene>. Therefore, the cytosolic cAc is named IREBP for IRE-binding protein when this function is talked about. Only one of the two functions is active, depending on whether <scene name='Aconitase/2b3x-cluster/1'>the (4Fe-4S) cofactor</scene> is present in the molecule: it's essential for <scene name='Aconitase/2b3x-total/1'>the ACO function</scene>. You can see, by <scene name='Aconitase/Morph/2'>looking at the morph</scene>, how much the enzyme structure differs between those two functions. |
Along with serving as a catalyst, aconitase is a member of the iron regulatory protien-1 (IRP-1) family. These enzymes have been found to play a role in regulatory RNA-binding proteins. This suggests a novel role for Fe-S clusters as post-translational regulatory switches.<ref name="Frishman" /> | Along with serving as a catalyst, aconitase is a member of the iron regulatory protien-1 (IRP-1) family. These enzymes have been found to play a role in regulatory RNA-binding proteins. This suggests a novel role for Fe-S clusters as post-translational regulatory switches.<ref name="Frishman" /> | ||
| + | |||
| + | == 3D structures of Aconitase== | ||
| + | [[Aconitase 3D structures]] | ||
| + | |||
</StructureSection> | </StructureSection> | ||
__NOTOC__ | __NOTOC__ | ||
| - | == 3D structures of Aconitase== | ||
| - | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| - | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| - | |||
| - | * ACO | ||
| - | |||
| - | **[[1b0k]] – pACO (mutant) – pig<br /> | ||
| - | **[[5acn]] – pACO+Fe3S4<br /> | ||
| - | **[[6acn]] - pACO+Fe4S4<br /> | ||
| - | **[[1amj]], [[1nit]] – cACO - cow<br /> | ||
| - | |||
| - | *ACO+citrate | ||
| - | |||
| - | **[[1c96]] - pACO (mutant)+citrate<br /> | ||
| - | **[[1b0m]] - pACO (mutant)+fluorocitrate<br /> | ||
| - | |||
| - | *ACO+aconitate | ||
| - | |||
| - | **[[1fgh]] – cACO+4-hydroxy-aconitate <br /> | ||
| - | **[[1aco]] – cACO+transaconitate<br /> | ||
| - | **[[1nis]] - cACO+transaconitate+nitrocitrate<br /> | ||
| - | |||
| - | *ACO+isocitrate | ||
| - | |||
| - | **[[7acn]] - pACO +isocitrate<br /> | ||
| - | **[[1c97]], [[1b0j]] - pACO (mutant)+isocitrate<br /> | ||
| - | **[[1ami]], [[8acn]] – cACO+isocitrate<br /> | ||
| - | |||
| - | * ACO1 | ||
| - | |||
| - | **[2b3x]], [[2b3y]] – hACO1 – human<br /> | ||
| - | **[[2ipy]], [[3snp]] – rACO1 (mutant)+ferritin H IRE-RNA – rabbit<br /> | ||
| - | **[[3sn2]] - rACO1 (mutant)+ transferrin receptor iron regulatory RNA<br /> | ||
| - | |||
| - | * ACO2 | ||
| - | **[[1l5j]] – ACO2 – ''Escherichia coli''<br /> | ||
| - | }} | ||
| - | <!--== Available structures == | ||
| - | In the PDB, nearly all deposited structures are from mammals, [[1l5j]] is from ''E.coli''. Also, only [[2ipy]] shows the IREBP function of cAc---it's also the only from rabbit. There are only two other cAc structures, with and without citrate, also the only from human. All other structures are either cow or pig, and a mutant from pig; all three proteins with several different ligands and inhibitors. | ||
| - | *[[1aco]] - mAc (''Bos taurus'') with ''trans''-aconitate (inhibitor) | ||
| - | *[[1ami]] - mAc (''Bos taurus'') with methylisocitrate | ||
| - | *[[1amj]] - mAc (''Bos taurus'') with sulfate and hydroxide | ||
| - | *[[1b0j]] - S642 mutant of mAc (''Sus scrofa'') with isocitrate (substrate) | ||
| - | *[[1b0k]] - S642 mutant of mAc (''Sus scrofa'') with fluorocitrate (inhibitor) | ||
| - | *[[1b0m]] - S642 mutant of mAc (''Sus scrofa'') with fluorocitrate (inhibitor) and oxygen | ||
| - | *[[1c96]] - S642 mutant of mAc (''Sus scrofa'') with citrate | ||
| - | *[[1c97]] - S642 mutant of mAc (''Sus scrofa'') with isocitrate and oxygen | ||
| - | *[[1fgh]] - mAc (''Bos taurus'') with 4-hydroxy-''trans''-aconitate (inhibitor) | ||
| - | *[[1l5j]] - aconitase B (''E. coli'') with Fe3S4 and aconitate | ||
| - | *[[1nis]] - mAc (''Bos taurus'') with nitrocitrate (inhibitor) | ||
| - | *[[1nit]] - mAc (''Bos taurus'') with sulfate | ||
| - | *[[2b3x]] - cAc (human) as aconitase with Fe4S4 | ||
| - | *[[2b3y]] - cAc (human) as aconitase with Fe4S4 and citrate | ||
| - | *[[2ipy]] - cAc (''Oryctolagus cuniculus'') as IRP1 with ferritin RNA | ||
| - | *[[5acn]] - mAc (''Sus scrofa'') with Fe3S4 (missing a Fe) | ||
| - | *[[6acn]] - mAc (''Sus scrofa'') with tricarballylic acid | ||
| - | *[[7acn]] - mAc (''Sus scrofa'') with isocitrate | ||
| - | *[[8acn]] - mAc (''Sus scrofa'') with nitroisocitrate | ||
| - | --> | ||
== Literature == | == Literature == | ||
* M. Claire Kennedy and Helmut Beinert: ''IX.4. Aconitase.'' in Ivano Bertini, Harry B. Gray, Edward I. Stiefel, Joan Selverstone Valentine (eds.): ''Biological Inorganic Chemistry: Structure and Reactivity.'' University Science Books, Herndon 2006. ISBN 1891389432 pp.209-- | * M. Claire Kennedy and Helmut Beinert: ''IX.4. Aconitase.'' in Ivano Bertini, Harry B. Gray, Edward I. Stiefel, Joan Selverstone Valentine (eds.): ''Biological Inorganic Chemistry: Structure and Reactivity.'' University Science Books, Herndon 2006. ISBN 1891389432 pp.209-- | ||
Current revision
| |||||||||||
Literature
- M. Claire Kennedy and Helmut Beinert: IX.4. Aconitase. in Ivano Bertini, Harry B. Gray, Edward I. Stiefel, Joan Selverstone Valentine (eds.): Biological Inorganic Chemistry: Structure and Reactivity. University Science Books, Herndon 2006. ISBN 1891389432 pp.209--
Additional Resources
For additional information, see: Carbohydrate Metabolism; Krebs cycle step 2.
References
- ↑ Zheng L, Kennedy MC, Beinert H, Zalkin H. Mutational analysis of active site residues in pig heart aconitase. J Biol Chem. 1992 Apr 15;267(11):7895-903. PMID:1313811
- ↑ 2.0 2.1 Frishman D, Hentze MW. Conservation of aconitase residues revealed by multiple sequence analysis. Implications for structure/function relationships. Eur J Biochem. 1996 Jul 1;239(1):197-200. PMID:8706708
- ↑ Dupuy J, Volbeda A, Carpentier P, Darnault C, Moulis JM, Fontecilla-Camps JC. Crystal structure of human iron regulatory protein 1 as cytosolic aconitase. Structure. 2006 Jan;14(1):129-39. PMID:16407072 doi:10.1016/j.str.2005.09.009
- ↑ 4.0 4.1 4.2 Beinert, H., Kennedy, M. C., Stout, C.D. “Aconitase as Iron−Sulfur Protein, Enzyme, and Iron-Regulatory Protein.” Chem. Rev. 1996, 96, 2335−2373.
- ↑ Lauble H, Kennedy MC, Beinert H, Stout CD. Crystal structures of aconitase with trans-aconitate and nitrocitrate bound. J Mol Biol. 1994 Apr 8;237(4):437-51. PMID:8151704 doi:http://dx.doi.org/10.1006/jmbi.1994.1246
- ↑ 6.0 6.1 6.2 6.3 Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry Life at the Molecular Level. New York: John Wiley & Sons, 2008. p. 578-579. Print.
- ↑ 7.0 7.1 Flint, DH., and Allen, RM. "Iron-sulfur protein with nonredox functions.” Chem. Rev. 1996, 96, 2315−2334.
External links
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Ralf Stephan, David Canner, Joel L. Sussman, Jaime Prilusky, Anthony Noles, Angel Herraez, Eran Hodis