This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sanbox glut3
From Proteopedia
(Difference between revisions)
| (One intermediate revision not shown.) | |||
| Line 1: | Line 1: | ||
==Facilitated Glucose Transporter 3, Solute Carrier Family 2 (GLUT3/ SLC2A3) in Homo Sapiens== | ==Facilitated Glucose Transporter 3, Solute Carrier Family 2 (GLUT3/ SLC2A3) in Homo Sapiens== | ||
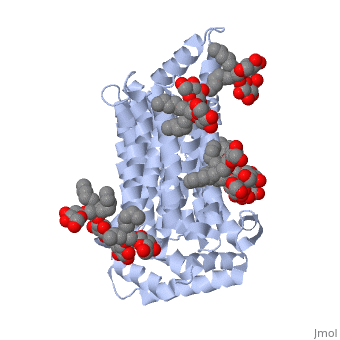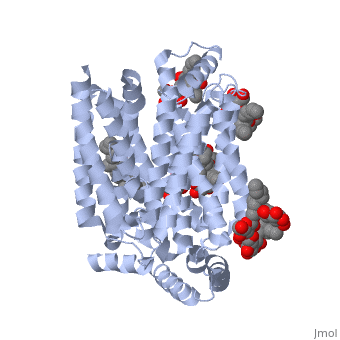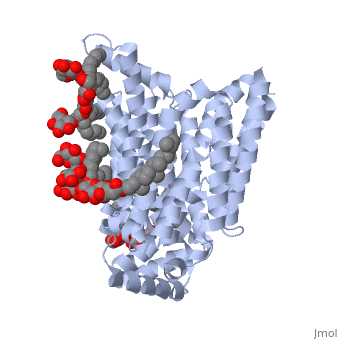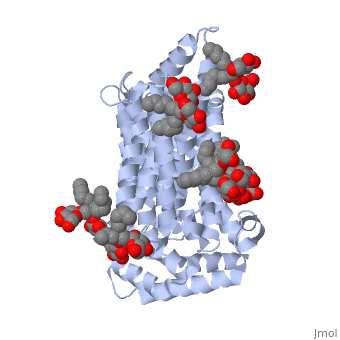
| - | <StructureSection load='5c65' size='340' side='right' caption=' | + | <StructureSection load='5c65' size='340' side='right' caption='Human sugar transporter complex with cholesterol hemisuccinate and octyl glucose neopentyl glycol (PDB code [[5c65]])'> |
== Function == | == Function == | ||
GLUT3 is one of fourteen facilitative sugar transporters, which use the glucose diffusion gradient to move across various plasma membranes to display various specificities, kinetics and tissue expression profiles <ref name="three">Long, W., & Cheeseman, C. I. (2015). Structure of, and functional insight into the GLUT family of membrane transporters. Cell Health and Cytoskeleton, 7, 167-183. doi:10.2147/CHC.S60484</ref>. Glucose transporters are approximately 500 amino acids in length and part of a growing superfamily of integral membrane glycoproteins that have 12 transmembrane (TM) helices. The transmembrane regions presumably create channels through which glucose can move<ref name="four">Kipmen-Korgun, D., Bilmen-Sarikcioglu, S., Altunbas, H., Demir, R., & Korgun, E. T. (2009). Type-2 diabetes down-regulates glucose transporter proteins and genes of the human blood leukocytes.Scandinavian Journal of Clinical and Laboratory Investigation, 69(3), 350-358. | GLUT3 is one of fourteen facilitative sugar transporters, which use the glucose diffusion gradient to move across various plasma membranes to display various specificities, kinetics and tissue expression profiles <ref name="three">Long, W., & Cheeseman, C. I. (2015). Structure of, and functional insight into the GLUT family of membrane transporters. Cell Health and Cytoskeleton, 7, 167-183. doi:10.2147/CHC.S60484</ref>. Glucose transporters are approximately 500 amino acids in length and part of a growing superfamily of integral membrane glycoproteins that have 12 transmembrane (TM) helices. The transmembrane regions presumably create channels through which glucose can move<ref name="four">Kipmen-Korgun, D., Bilmen-Sarikcioglu, S., Altunbas, H., Demir, R., & Korgun, E. T. (2009). Type-2 diabetes down-regulates glucose transporter proteins and genes of the human blood leukocytes.Scandinavian Journal of Clinical and Laboratory Investigation, 69(3), 350-358. | ||
| Line 7: | Line 7: | ||
==Structure== | ==Structure== | ||
GLUT3(<scene name='71/716527/5c65/1'>5c65</scene>) is a transport protein consisting of 481 amino acids and weighing 52,520 Daltons in its asymmetrical unit<ref name="nineteen">http://www.ebi.ac.uk/pdbe/entry/pdb/5c65/</ref>. This protein is an alpha-helical protein consisting of two chains, two different ligands and water<ref name="nineteen"/>. The structure was determined by X-Ray diffraction and was measured at a resolution of 2.65 Angstroms<ref name="twentytwo">http://oca.weizmann.ac.il/oca-bin/ocaids?id=5c65</ref>. GLUT3 consists of 12 transmembrane segments (TMs) folded “into the N-terminal and C-terminal domains, each comprising ‘3+3’ inverted repeats”<ref name="nine"/> These TMs consist of four 3 repeated sections. [http://www.nature.com/nature/journal/v526/n7573/fig_tab/nature14655_F1.html Here] is a figure by Deng, D., et al. showing these repeated transmembrane segments<ref name="nine">Deng, D., Sun, P., Yan, C., Ke, M., Jiang, X., Xiong, L., . . . Yan, N. (2015). Molecular basis of ligand recognition and transport by glucose transporters. Nature, 526(7573), 391-396. doi:10.1038/nature14655</ref>. The protein consists of two different ligands, Y01 and 37X<ref name="eighteen">http://www.rcsb.org/pdb/explore.do?structureId=5C65</ref>. Octyl Glucose Neopentyl Glycol (<scene name='71/716527/37x/5'>37X</scene>) has a chemical formula of C<sub>27</sub>H<sub>52</sub>O<sub>12</sub> and a molecular weight of 569 Da. There are six 37X (501-506a) bound to chain A of 5c65. These ligands are kept in place by hydrogen bonds to arginine, proline, and serine and by van der Waals forces. Chain B has three 37X ligands attached to it (501-503b). These are attached through hydrogen bonds by arginine, proline, and serine as well as by van der Waals forces<ref name="twenty">http://www.ebi.ac.uk/pdbe/entry/pdb/5c65/bound/37X</ref>. To view 37X in 3D use [http://www.rcsb.org/pdb/explore/jmol.do?structureId=5C65&residueNr=37X JSmol]. Cholesterol hemisuccinate (<scene name='71/716527/Y01/3'>Y01</scene>) has a chemical formula of C<sub>31</sub>H<sub>50</sub>O<sub>4</sub> and has a molecular weight of 487 Da. One Y01 is attached to chain a and another Y01 is attached to chain b<ref name="twentyone">http://www.ebi.ac.uk/pdbe/entry/pdb/5c65/bound/Y01</ref>. To view Y01 in 3D use [http://www.rcsb.org/pdb/explore/jmol.do?structureId=5C65&residueNr=Y01 JSmol]. GLUT3 was also identified and analyzed in a complex with alpha & beta d-glucose. This model was reported with a resolution of 1.5 Å and was in an open-occluded state<ref name="nine"/>. The alpha and beta d glucose were coordinated in a <scene name='71/716527/Binding_pocket/1'>binding pocket</scene> by amino acids N315, E378, Q159, W368, Q280, Q281, N286. These are located on TM8 and TM10a and TM10b<ref name="nine"/>. A figure of this glucose coordination by Deng, D., et al. is available [http://www.nature.com/nature/journal/v526/n7573/fig_tab/nature14655_F2.html here]. GLUT3 structure was also determined when bound to maltose in an outward-open and an outward-occluded conformation. This was measure to a resolution of 2.6 Å and 2.4 Å respectively. A figure of this maltose coordination by Deng, D., et al. is available [http://www.nature.com/nature/journal/v526/n7573/fig_tab/nature14655_F2.html here]. To get a better view of the structure of the protein use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=5C65 FirstGlance]. | GLUT3(<scene name='71/716527/5c65/1'>5c65</scene>) is a transport protein consisting of 481 amino acids and weighing 52,520 Daltons in its asymmetrical unit<ref name="nineteen">http://www.ebi.ac.uk/pdbe/entry/pdb/5c65/</ref>. This protein is an alpha-helical protein consisting of two chains, two different ligands and water<ref name="nineteen"/>. The structure was determined by X-Ray diffraction and was measured at a resolution of 2.65 Angstroms<ref name="twentytwo">http://oca.weizmann.ac.il/oca-bin/ocaids?id=5c65</ref>. GLUT3 consists of 12 transmembrane segments (TMs) folded “into the N-terminal and C-terminal domains, each comprising ‘3+3’ inverted repeats”<ref name="nine"/> These TMs consist of four 3 repeated sections. [http://www.nature.com/nature/journal/v526/n7573/fig_tab/nature14655_F1.html Here] is a figure by Deng, D., et al. showing these repeated transmembrane segments<ref name="nine">Deng, D., Sun, P., Yan, C., Ke, M., Jiang, X., Xiong, L., . . . Yan, N. (2015). Molecular basis of ligand recognition and transport by glucose transporters. Nature, 526(7573), 391-396. doi:10.1038/nature14655</ref>. The protein consists of two different ligands, Y01 and 37X<ref name="eighteen">http://www.rcsb.org/pdb/explore.do?structureId=5C65</ref>. Octyl Glucose Neopentyl Glycol (<scene name='71/716527/37x/5'>37X</scene>) has a chemical formula of C<sub>27</sub>H<sub>52</sub>O<sub>12</sub> and a molecular weight of 569 Da. There are six 37X (501-506a) bound to chain A of 5c65. These ligands are kept in place by hydrogen bonds to arginine, proline, and serine and by van der Waals forces. Chain B has three 37X ligands attached to it (501-503b). These are attached through hydrogen bonds by arginine, proline, and serine as well as by van der Waals forces<ref name="twenty">http://www.ebi.ac.uk/pdbe/entry/pdb/5c65/bound/37X</ref>. To view 37X in 3D use [http://www.rcsb.org/pdb/explore/jmol.do?structureId=5C65&residueNr=37X JSmol]. Cholesterol hemisuccinate (<scene name='71/716527/Y01/3'>Y01</scene>) has a chemical formula of C<sub>31</sub>H<sub>50</sub>O<sub>4</sub> and has a molecular weight of 487 Da. One Y01 is attached to chain a and another Y01 is attached to chain b<ref name="twentyone">http://www.ebi.ac.uk/pdbe/entry/pdb/5c65/bound/Y01</ref>. To view Y01 in 3D use [http://www.rcsb.org/pdb/explore/jmol.do?structureId=5C65&residueNr=Y01 JSmol]. GLUT3 was also identified and analyzed in a complex with alpha & beta d-glucose. This model was reported with a resolution of 1.5 Å and was in an open-occluded state<ref name="nine"/>. The alpha and beta d glucose were coordinated in a <scene name='71/716527/Binding_pocket/1'>binding pocket</scene> by amino acids N315, E378, Q159, W368, Q280, Q281, N286. These are located on TM8 and TM10a and TM10b<ref name="nine"/>. A figure of this glucose coordination by Deng, D., et al. is available [http://www.nature.com/nature/journal/v526/n7573/fig_tab/nature14655_F2.html here]. GLUT3 structure was also determined when bound to maltose in an outward-open and an outward-occluded conformation. This was measure to a resolution of 2.6 Å and 2.4 Å respectively. A figure of this maltose coordination by Deng, D., et al. is available [http://www.nature.com/nature/journal/v526/n7573/fig_tab/nature14655_F2.html here]. To get a better view of the structure of the protein use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=5C65 FirstGlance]. | ||
| - | [[Image:http://www.nature.com/nature/journal/v526/n7573/images/nature14655-f2.jpg]] | ||
This is 5c65 shown with <scene name="/12/3456/Sample/1">colored groups</scene>. This is 5c65 shown as a <scene name="/12/3456/Sample/2">transparent representation</scene> of the protein. | This is 5c65 shown with <scene name="/12/3456/Sample/1">colored groups</scene>. This is 5c65 shown as a <scene name="/12/3456/Sample/2">transparent representation</scene> of the protein. | ||
Current revision
Facilitated Glucose Transporter 3, Solute Carrier Family 2 (GLUT3/ SLC2A3) in Homo Sapiens
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 Long, W., & Cheeseman, C. I. (2015). Structure of, and functional insight into the GLUT family of membrane transporters. Cell Health and Cytoskeleton, 7, 167-183. doi:10.2147/CHC.S60484
- ↑ 2.0 2.1 Kipmen-Korgun, D., Bilmen-Sarikcioglu, S., Altunbas, H., Demir, R., & Korgun, E. T. (2009). Type-2 diabetes down-regulates glucose transporter proteins and genes of the human blood leukocytes.Scandinavian Journal of Clinical and Laboratory Investigation, 69(3), 350-358. doi:10.1080/00365510802632163
- ↑ 3.0 3.1 Simpson,I. A., Dwyer, D., Malide, D., Moley, K. H., Travis, A., & Vannucci, S. J. (2008). The facilitative glucose transporter GLUT3: 20 years of distinction. American Journal of Physiology - Endocrinology and Metabolism, 295(2), E242-E253. doi:10.1152/ajpendo.90388.2008
- ↑ Maher, F., Vannucci, S. J., & Simpson, I. A. (1994). Glucose transporter proteins in brain. FASEB Journal, 8(13), 1003-1011.
- ↑ Xu, J., Lu, C., Wang, J., Zhang, R., Qian, X., & Zhu, H. (2015). Regulation of human trophoblast GLUT3 glucose transporter by mammalian target of rapamycin signaling. International Journal of Molecular Sciences, 16(6), 13815-13828. doi:10.3390/ijms160613815
- ↑ 6.0 6.1 Liu, Y., Liu, F., Iqbal, K., Grundke-Iqbal, I., & Gong, C. -. (2008). Decreased glucose transporters correlate to abnormal hyperphosphorylation of tau in alzheimer disease. FEBS Letters, 582(2), 359-364. doi:10.1016/j.febslet.2007.12.035
- ↑ 7.0 7.1 http://www.ebi.ac.uk/pdbe/entry/pdb/5c65/
- ↑ http://oca.weizmann.ac.il/oca-bin/ocaids?id=5c65
- ↑ 9.0 9.1 9.2 9.3 Deng, D., Sun, P., Yan, C., Ke, M., Jiang, X., Xiong, L., . . . Yan, N. (2015). Molecular basis of ligand recognition and transport by glucose transporters. Nature, 526(7573), 391-396. doi:10.1038/nature14655
- ↑ http://www.rcsb.org/pdb/explore.do?structureId=5C65
- ↑ http://www.ebi.ac.uk/pdbe/entry/pdb/5c65/bound/37X
- ↑ http://www.ebi.ac.uk/pdbe/entry/pdb/5c65/bound/Y01
- ↑ Vittori, A., Breda, C., Repici, M., Orth, M., Roos, R. A. C., Outeiro, T. F., . . . the REGISTRY investigators of the European Huntington's Disease Network. (2014). Copy-number variation of the neuronal glucose transporter gene SLC2A3 and age of onset in huntington's disease. Human Molecular Genetics, 23(12), 3129-3137. doi:10.1093/hmg/ddu022
- ↑ McClory, H., Williams, D., & Sapp, E. (2014). Glucose transporter 3 is a rab11-dependent trafficking cargo and its transport to the cell surface is reduced in neurons of CAG140 Huntington’s disease mice. Acta Neuropathol Commun, 2, 1-9.
- ↑ 15.0 15.1 Naftalin RJ, Holman GD. Transport of sugars in human red cells. In: Ellory JC, Lew V, editors. \ Membrane Transport in Red Cells. New York, NY, USA: Academic Press; 1977.
- ↑ 16.0 16.1 Carruthers, A., DeZutter, J., Ganguly, A., & Devaskar, S. U. (2009). Will the original glucose transporter isoform please stand up! American Journal of Physiology - Endocrinology and Metabolism, 297(4), E836-E848. doi:10.1152/ajpendo.00496.2009
- ↑ Jardetzky, O. (1966). Simple allosteric model for membrane pumps [27]. Nature, 211(5052), 969-970. doi:10.1038/211969a0
- ↑ Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615.
- ↑ Caulfield MJ, Munroe PB, O’Neill D, et al. SLC2A9 is a high-capacity urate transporter in humans. PLoS Med. 2008;5:1509–1523.
- ↑ Vollers, S. S., & Carruthers, A. (2012). Sequence determinants of GLUT1-mediated accelerated-exchange transport: Analysis by homology-scanning mutagenesis. Journal of Biological Chemistry, 287(51), 42533-42544.doi:10.1074/jbc.M112.369587