Chymotrypsin
From Proteopedia
| (15 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
<StructureSection load='7gch' size='350' side='right' scene='38/387136/Bovine_chymotrypsin_overview/1' caption='Bovine γ-chymotrypsin A (residues 1-13 in pink, 16-146 in cyan, 149-245 in gold) complex with inhibitor (PDB code [[7gch]]) '> | <StructureSection load='7gch' size='350' side='right' scene='38/387136/Bovine_chymotrypsin_overview/1' caption='Bovine γ-chymotrypsin A (residues 1-13 in pink, 16-146 in cyan, 149-245 in gold) complex with inhibitor (PDB code [[7gch]]) '> | ||
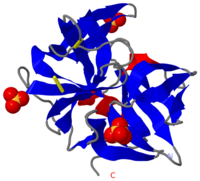
[[Image:2ea3.png|left|200px|thumb|Crystal Structure of ''Cellulomonas Bogoriensis'' Chymotrypsin [[2ea3]]]] | [[Image:2ea3.png|left|200px|thumb|Crystal Structure of ''Cellulomonas Bogoriensis'' Chymotrypsin [[2ea3]]]] | ||
| - | [[Chymotrypsin]] (Chy or α-Chy) is a digestive enzyme containing an active serine residue, which helps to digest proteins in our food. Other related proteases are crucial for blood clotting ([http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=stryer&part=A1378&rendertype=figure&id=A1401 thrombin and other proteases]), for the AIDS virus metabolism ([http://www.proteopedia.org/wiki/index.php/Hiv_protease HIV protease]) and for many other processes relevant to human health and agriculture. Chymotrypsin cleaves peptide bonds of proteins where the amide side of the bond is an aromatic amino acid like tyrosine, phenylalanine or tryptophan. Bovine Chy is found in 2 forms: A and B. The 2 forms have different proteolytic characteristics | + | __TOC__ |
| + | == Function == | ||
| + | [[Chymotrypsin]] (Chy or α-Chy) is a digestive enzyme containing an active serine residue, which helps to digest proteins in our food. Other related proteases are crucial for blood clotting ([http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=stryer&part=A1378&rendertype=figure&id=A1401 thrombin and other proteases]), for the AIDS virus metabolism ([http://www.proteopedia.org/wiki/index.php/Hiv_protease HIV protease]) and for many other processes relevant to human health and agriculture. Chymotrypsin cleaves peptide bonds of proteins where the amide side of the bond is an aromatic amino acid like tyrosine, phenylalanine or tryptophan. Bovine Chy is found in 2 forms: A and B. The 2 forms have different proteolytic characteristics. The image at the left is the crystal structure of chymotrypsin from ''Cellulomonas Bogoriensis'' ([[2ea3]]) with sulfate ions.<ref>PMID:3555886</ref><br /> | ||
| + | *'''γ-Chy''' is a covalent acyl adduct of α-Chy. | ||
| + | *'''Chymotrypsinogen''' is nonactive precursor of α-Chy. It is converted to the active form, i.e., chymotrypsin by trypsin. | ||
| + | Some additional details in<br /> | ||
*[[Molecular Playground/Chymotrypsin]]<br /> | *[[Molecular Playground/Chymotrypsin]]<br /> | ||
| + | *[[Protease]]<br /> | ||
*[[Serine Proteases]]. | *[[Serine Proteases]]. | ||
| Line 16: | Line 22: | ||
This view shows the <scene name='38/387136/Bovine_chymotrypsin_active_sit/3'>carbonyl group of the inhibitor</scene> in CPK colors. The triflouromethyl group is bound to the carbonyl carbon via the yellow bond. In a peptide substrate, the triflouromethyl group would be replaced by the first amino acid residue of the rest of the peptide chain, and the yellow bond would be the bond that is cleaved. The carbonyl carbon of the inhibitor is 1.52 Å away from the side chain oxygen of serine 195, and this indicates they are covalently bound (bond indicated by dotted line). Thus, this structure is similar to the '''tetrahedral intermediate''' that is formed during the cleavage reaction. The negative charge that develops on the carbonyl oxygen of the substrate is stabilized by hydrogen bonds to the backbone nitrogens of Ser 195 and Gly 193, shown in blue spacefill. The hydrogen atoms involved in these hydrogen bonds are not shown. | This view shows the <scene name='38/387136/Bovine_chymotrypsin_active_sit/3'>carbonyl group of the inhibitor</scene> in CPK colors. The triflouromethyl group is bound to the carbonyl carbon via the yellow bond. In a peptide substrate, the triflouromethyl group would be replaced by the first amino acid residue of the rest of the peptide chain, and the yellow bond would be the bond that is cleaved. The carbonyl carbon of the inhibitor is 1.52 Å away from the side chain oxygen of serine 195, and this indicates they are covalently bound (bond indicated by dotted line). Thus, this structure is similar to the '''tetrahedral intermediate''' that is formed during the cleavage reaction. The negative charge that develops on the carbonyl oxygen of the substrate is stabilized by hydrogen bonds to the backbone nitrogens of Ser 195 and Gly 193, shown in blue spacefill. The hydrogen atoms involved in these hydrogen bonds are not shown. | ||
| - | </StructureSection> | ||
| - | == 3D Structures of Chymotrypsin == | ||
| - | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| - | + | </StructureSection> | |
| - | + | ||
| - | + | == Mechanism == | |
| + | The hydrolysis occurs in two steps. First, the peptide bond with the C-terminal part of the substrate is replaced by an ester bond to the active site serine. Second, the covalent intermediate is hydrolyzed by water, releasing the N-terminal part of the substrate as carboxylic acid. Both steps occur via a tetrahedral intermediate containing an oxyanion that is stabilized by hydrogen bond donors lining the so-called oxyanion hole. Throughout the reaction, active site residue histidine 57 acts as base or acid to deprotonate nucleophiles or protonate leaving groups, respectively. The following 5-minute video shows the mechanism step by step. | ||
| - | + | <center><html5media height=“720” width=“1280” frameborder="0" allowfullscreen>https://www.youtube.com/embed/JWtjYYl5Ylw</html5media></center> | |
| - | + | <center>Mechanism of chymotrypsin ([http://proteopedia.org/wiki/index.php/Image:Chymotrypsin_mechanism.gz image source])</center> | |
| - | + | ||
| - | + | ||
| - | + | == 3D Structures of Chymotrypsin == | |
| - | + | [[Chymotrypsin 3D structures]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | **[[2cga]], [[1chg]], [[1ex3]] – bChygenA<br /> | ||
| - | **[[2jet]] – rChygenB chain A,B <br /> | ||
| - | + | == References == | |
| + | <references/> | ||
| - | **[[1gl0]], [[1gli]] – ChygenA+PMP_D2v – ''Locusta migratoria''<br /> | ||
| - | **[[1k2i]] - bChygenA+7-hydroxycoumarin<br /> | ||
| - | **[[2y6t]] – bChygenA + ecotin<br /> | ||
| - | **[[3t62]] - bChygenA + Kunitz-type proteinase inhibitor SHPI-1<br /> | ||
| - | **[[1cgi]], [[1cgj]] - bChygenA + Kazal-type inhibitor<br /> | ||
| - | **[[3ru4]] - bChygenA + trypsin + Bowman-Birk inhibitor<br /> | ||
| - | **[[1pyt]] – bChygenC + procarboxipeptidase + proproteinase<br /> | ||
| - | }} | ||
==Further reading== | ==Further reading== | ||
You can learn more about chymotrypsin structure, function and regulation in this publicly available [http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=stryer&part=A1170#A1171 chapter] of the Biochemistry textbook by Berg, Tymoczka and Stryer. | You can learn more about chymotrypsin structure, function and regulation in this publicly available [http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=stryer&part=A1170#A1171 chapter] of the Biochemistry textbook by Berg, Tymoczka and Stryer. | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
Contents |
Mechanism
The hydrolysis occurs in two steps. First, the peptide bond with the C-terminal part of the substrate is replaced by an ester bond to the active site serine. Second, the covalent intermediate is hydrolyzed by water, releasing the N-terminal part of the substrate as carboxylic acid. Both steps occur via a tetrahedral intermediate containing an oxyanion that is stabilized by hydrogen bond donors lining the so-called oxyanion hole. Throughout the reaction, active site residue histidine 57 acts as base or acid to deprotonate nucleophiles or protonate leaving groups, respectively. The following 5-minute video shows the mechanism step by step.
3D Structures of Chymotrypsin
References
- ↑ Appel W. Chymotrypsin: molecular and catalytic properties. Clin Biochem. 1986 Dec;19(6):317-22. PMID:3555886
Further reading
You can learn more about chymotrypsin structure, function and regulation in this publicly available chapter of the Biochemistry textbook by Berg, Tymoczka and Stryer.
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Karsten Theis, Alice Harmon, Alexander Berchansky