Sandbox flippases
From Proteopedia
(Difference between revisions)
| (3 intermediate revisions not shown.) | |||
| Line 3: | Line 3: | ||
==Flippases== | ==Flippases== | ||
<StructureSection load='5c76' size='340' side='right' caption='Caption for this structure' scene=''> | <StructureSection load='5c76' size='340' side='right' caption='Caption for this structure' scene=''> | ||
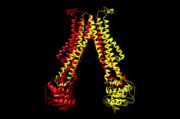
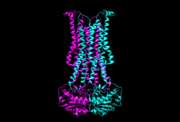
| - | '''Flippases''' are the homodimeric transmembrane proteins located in the membrane. It helps phospholipid to transport from inner | + | '''Flippases''' are the homodimeric transmembrane proteins located in the membrane. It helps phospholipid to transport from inner leaflet to outer leaflet of the cell membrane. It is composed with alpha-helix and beta sheets. The main three structural features of each monomer are an external helix, a belt of positively charged amino acids, and a binding site for an ATP molecule. |
| - | + | ||
| - | + | ||
== Structure == | == Structure == | ||
| - | Flippase is homodimer which is quartinary strucure. Each monomer is tertiary structure composed with nine alpha-helices and five beta sheets. The secondary structure, helix is composed with amino acids which are primary structure. The main three structural features of each monomer are an external helix, a belt of positively charged amino acids, and a binding site for an ATP molecule. External helix is hydrophobic groove and it is composed with amino acids(K55,Y63,Y56,R53,D47,Y50,T43). A positively charged belt has four Arginines(R302,R86,R260,R309) which are negative charged. | + | Flippase is homodimer which is quartinary strucure. Each monomer is tertiary structure composed with nine alpha-helices and five beta sheets. The secondary structure, helix is composed with amino acids which are primary structure. The main three structural features of each monomer are an external helix, a belt of positively charged amino acids, and a binding site for an ATP molecule. External helix is hydrophobic groove and it is composed with amino acids(K55,Y63,Y56,R53,D47,Y50,T43). It is located at the top, outer(periplasmic) side. A positively charged belt has four Arginines(R302,R86,R260,R309) which are negative charged. Binding site for an ATP molecule is located at the bottom, on the inner(cytoplasmic) side. As the ATP is bound, the flipping processing is began. |
== Function == | == Function == | ||
| - | + | As the tail of LLO is attached to external helix, it acts as the switch for the ATPase to supply the ATP. The membrane protein PglK used energy given by ATP hydrolysis to flip the lipid(LLO) from cytoplasmic(in) side to periplasmic(out) side of the membrane. The positively charged belt is placed in the inside of the outward-facing cavity. The belt region attract the pyrophosphate in the LLO headgroup which is negatively charged. The fold of PglK is similar to other ABC exporters. But PglK has an external helix (short alpha-helix) parallel to the periplasmic side <ref> Perez, C., Gerber, S., Boilevin, J., Bucher, M., Darbre, T., Aebi, M., ... & Locher, K. P. (2015). Structure and mechanism of an active lipid-linked oligosaccharide flippase. Nature, 524(7566), 433-438. </ref>. | |
| - | == Mechanism == | + | == Mechanism of PglK == |
The beginning conformation is an outward-occluded conformation and the head of Linked-lipid Oligosaccharide(LLO) is on the cytoplasmic side. When the tail of LLO is attached to external helix, the ADP exchanges to ATP and the conformation changes to outward-open state. The head of LLO attaches to a positively charged belt facing to periplasmic side since the pyrophosphate has negative charge which attracts to positive charge. As ATP changes to ADP, the conformation returns to outward-occluded conformation and it squeezes the LLO head out to periplasmic side of the membrane and the tail of LLO is released to extend to cytoplasmic side. Therefore, the position of LLO is opposite through this mechanism. <ref name= "name"> Verchère, A., & Menon, A. K. (2015). Structural biology: Lipid gymnastics. Nature, 524(7566), 420-422. </ref> | The beginning conformation is an outward-occluded conformation and the head of Linked-lipid Oligosaccharide(LLO) is on the cytoplasmic side. When the tail of LLO is attached to external helix, the ADP exchanges to ATP and the conformation changes to outward-open state. The head of LLO attaches to a positively charged belt facing to periplasmic side since the pyrophosphate has negative charge which attracts to positive charge. As ATP changes to ADP, the conformation returns to outward-occluded conformation and it squeezes the LLO head out to periplasmic side of the membrane and the tail of LLO is released to extend to cytoplasmic side. Therefore, the position of LLO is opposite through this mechanism. <ref name= "name"> Verchère, A., & Menon, A. K. (2015). Structural biology: Lipid gymnastics. Nature, 524(7566), 420-422. </ref> | ||
[[Image:5c76.png | thumb | 5c76, outward-open conformation]] | [[Image:5c76.png | thumb | 5c76, outward-open conformation]] | ||
[[Image:5c73.png | thumb | 5c73, outward-occluded conformation]] | [[Image:5c73.png | thumb | 5c73, outward-occluded conformation]] | ||
| - | + | ||
| - | + | ||
== Disease == | == Disease == | ||
| - | + | The loss of asymmetry could cause apoptosis. This could affect to neurons which cause Alzheimer's disease by neurodegenerative. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | This | + | |
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Current revision
This page is setup for Leah to build her senior project for OU CHEM 4923
Flippases
| |||||||||||
References
- ↑ Perez, C., Gerber, S., Boilevin, J., Bucher, M., Darbre, T., Aebi, M., ... & Locher, K. P. (2015). Structure and mechanism of an active lipid-linked oligosaccharide flippase. Nature, 524(7566), 433-438.
- ↑ Verchère, A., & Menon, A. K. (2015). Structural biology: Lipid gymnastics. Nature, 524(7566), 420-422.